Recent from talks
Nothing was collected or created yet.
Longest common substring
View on WikipediaIn computer science, a longest common substring of two or more strings is a longest string that is a substring of all of them. There may be more than one longest common substring. Applications include data deduplication and plagiarism detection.
Unlike the longest common subsequence problem, which finds insertions or deletions within the common text, the longest common substring problem seeks a contiguous substring shared by both texts.
Examples
[edit]
The picture shows two strings where the problem has multiple solutions. Although the substring occurrences always overlap, it is impossible to obtain a longer common substring by "uniting" them.
The strings "ABABC", "BABCA" and "ABCBA" have only one longest common substring, viz. "ABC" of length 3. Other common substrings are "A", "AB", "B", "BA", "BC" and "C".
ABABC
|||
BABCA
|||
ABCBA
Problem definition
[edit]Given two strings, of length and of length , find a longest string which is substring of both and .
A generalization is the k-common substring problem. Given the set of strings , where and . Find for each , a longest string which occurs as substring of at least strings.
Algorithms
[edit]One can find the lengths and starting positions of the longest common substrings of and in time with the help of a generalized suffix tree. A faster algorithm can be achieved in the word RAM model of computation if the size of the input alphabet is in . In particular, this algorithm runs in time using space.[1] Solving the problem by dynamic programming costs . The solutions to the generalized problem take space and time with dynamic programming and take time with a generalized suffix tree.
Suffix tree
[edit]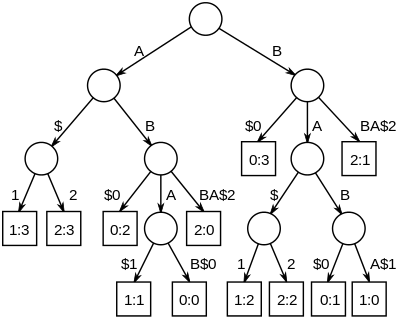
The longest common substrings of a set of strings can be found by building a generalized suffix tree for the strings, and then finding the deepest internal nodes which have leaf nodes from all the strings in the subtree below it. The figure on the right is the suffix tree for the strings "ABAB", "BABA" and "ABBA", padded with unique string terminators, to become "ABAB$0", "BABA$1" and "ABBA$2". The nodes representing "A", "B", "AB" and "BA" all have descendant leaves from all of the strings, numbered 0, 1 and 2.
Building the suffix tree takes time (if the size of the alphabet is constant). If the tree is traversed from the bottom up with a bit vector telling which strings are seen below each node, the k-common substring problem can be solved in time. If the suffix tree is prepared for constant time lowest common ancestor retrieval, it can be solved in time.[2]
Dynamic programming
[edit]The following pseudocode finds the set of longest common substrings between two strings with dynamic programming:
function LongestCommonSubstring(S[1..r], T[1..n])
L := array(1..r, 1..n)
z := 0 # length of the longest common substring found so far
ret := {}
for i := 1..r
for j := 1..n
if S[i] = T[j]
if i = 1 or j = 1
L[i, j] := 1
else
L[i, j] := L[i − 1, j − 1] + 1
if L[i, j] > z
z := L[i, j]
ret := {S[(i − z + 1)..i]}
else if L[i, j] = z
ret := ret ∪ {S[(i − z + 1)..i]}
else
L[i, j] := 0
return ret
This algorithm runs in time. The array L stores the length of the longest common suffix of the prefixes S[1..i] and T[1..j] which end at position i and j, respectively. The variable z is used to hold the length of the longest common substring found so far. The set ret is used to hold the set of strings which are of length z. The set ret can be saved efficiently by just storing the index i, which is the last character of the longest common substring (of size z) instead of S[(i-z+1)..i]. Thus all the longest common substrings would be, for each i in ret, S[(ret[i]-z)..(ret[i])].
The following tricks can be used to reduce the memory usage of an implementation:
- Keep only the last and current row of the DP table to save memory ( instead of )
- The last and current row can be stored on the same 1D array by traversing the inner loop backwards
- Store only non-zero values in the rows. This can be done using hash-tables instead of arrays. This is useful for large alphabets.
See also
[edit]- Longest palindromic substring
- n-gram, all the possible substrings of length n that are contained in a string
References
[edit]- ^ Charalampopoulos, Panagiotis; Kociumaka, Tomasz; Pissis, Solon P.; Radoszewski, Jakub (Aug 2021). Mutzel, Petra; Pagh, Rasmus; Herman, Grzegorz (eds.). Faster Algorithms for Longest Common Substring. European Symposium on Algorithms. Leibniz International Proceedings in Informatics (LIPIcs). Vol. 204. Schloss Dagstuhl. doi:10.4230/LIPIcs.ESA.2021.30. Here: Theorem 1, p.30:2.
- ^ Gusfield, Dan (1999) [1997]. Algorithms on Strings, Trees and Sequences: Computer Science and Computational Biology. USA: Cambridge University Press. ISBN 0-521-58519-8.
External links
[edit]- Dictionary of Algorithms and Data Structures: longest common substring
- Perl/XS implementation of the dynamic programming algorithm
- Perl/XS implementation of the suffix tree algorithm
- Dynamic programming implementations in various languages on wikibooks
- working AS3 implementation of the dynamic programming algorithm
- Suffix Tree based C implementation of Longest common substring for two strings
Longest common substring
View on GrokipediaFundamentals
Formal Definition
A substring of a string is defined as a contiguous sequence of characters within that string. The longest common substring problem, given two strings and over an alphabet , consists of finding the longest string that is a substring of both and , along with its length and the starting positions where it occurs in and .[4] The notation is commonly used to denote this longest string , while refers to its length.[4] Special cases arise depending on the input strings. If either or is the empty string, then is the empty string with length 0.[5] If and are identical, then , so the length equals . If and share no common non-empty substring (e.g., consisting of disjoint alphabets), the length is 0, corresponding to the empty string.[5] The problem generalizes to strings , where the goal is to find the longest string that is a substring of every for to , again including its length and positions in each string.[6] In this setting, denotes the resulting string, and the empty string serves as the trivial solution with length 0 when no longer common substring exists.[6] This generalization can be computed using methods such as dynamic programming extended to multiple dimensions.[6]Key Properties
The longest common substring (LCS) of two strings is not necessarily unique, as multiple distinct substrings of the maximum length may occur in both strings simultaneously. In practice, identifying any one such substring suffices for most applications, since the problem typically seeks the length or a representative instance rather than an exhaustive enumeration. A defining characteristic of the LCS is its contiguous nature, requiring the common characters to appear consecutively in both input strings without gaps. This distinguishes it from the longest common subsequence, where elements need not be adjacent, allowing for more flexible but computationally harder alignments in sequence comparison tasks. The contiguity constraint simplifies certain structural analyses, such as those using suffix-based representations. Occurrences of the LCS within the input strings can overlap, meaning the same maximal substring may appear in multiple positions that share characters. This overlapping property arises naturally in strings with repetitive patterns and influences how multiple instances are detected in advanced string processing. The LCS length demonstrates monotonicity: if string U is a substring of string V, then for any string T, the LCS length between U and T is at most the LCS length between V and T. This follows directly from the substring inclusion, as every common substring of U and T remains a valid common substring of V and T. The LCS length remains invariant under certain transformations, such as reversing both input strings; the length of the LCS between S and T equals that between the reverses of S and T. This invariance holds because reversal preserves the relative order and adjacency of characters within substrings, merely flipping their orientation without altering lengths.Examples and Illustrations
Basic Examples with Two Strings
Consider two strings S = "ABAB" and T = "BABA". To identify the longest common substring manually, examine contiguous sequences starting from each position in S and check for matches in T. For instance, starting at position 1 in S ("ABA"), this sequence matches positions 2 through 4 in T ("ABA"). Similarly, starting at position 2 in S ("BAB"), it matches positions 1 through 3 in T ("BAB"). Each of these substrings has length 3, establishing the maximum length for common contiguous segments in this pair. Shorter common substrings, such as "AB" (length 2, appearing multiple times in both), are discarded because the problem requires the longest possible match. The following alignment visualizes one such longest common substring ("ABA"):S: A B A B
↑ ↑ ↑
T: B A B A
↑ ↑ ↑
S: A B A B
↑ ↑ ↑
T: B A B A
↑ ↑ ↑
| S Positions | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| S | a | b | c | d | x | y | z |
| T Positions | 4 | 5 | 6 | 7 | |||
| T | a | b | c | d |
Examples with Multiple Strings
When extending the longest common substring problem to multiple strings, the goal is to find the longest contiguous sequence that appears in every string in the set, representing the intersection of common substrings across all inputs. This generalization is particularly useful in computational biology for identifying conserved motifs in sets of DNA or protein sequences from related organisms. The longest common substring length typically decreases as more strings are added, since the substring must satisfy the consecutive matching condition in each one. Consider three strings: S1 = "BANANA", S2 = "BAYAN", and S3 = "BABA". The longest common substring to all three is "BA", with length 2. To illustrate the distinction from pairwise comparisons, note that the pairwise longest common substring lengths are also 2: "BA" or "AN" between S1 and S2, "BA" between S1 and S3, and "BA" between S2 and S3. However, no substring of length 3 is shared by all three, highlighting how the multi-string case requires stricter intersection. A similar example arises in DNA sequence analysis, where short strings represent segments of genetic material. For S1 = "ABABC", S2 = "BABCA", and S3 = "ABCBA" (using letters analogous to nucleotide bases A, B, C for simplicity), the longest common substring is "ABC", with length 3. Here, pairwise longest common substrings can be longer—for instance, "BABC" (length 4) between S1 and S2—but the overall longest common substring shortens to 3 when intersecting with S3. Such patterns help identify evolutionarily conserved regions across multiple genomes. Adding more strings often reduces the longest common substring length, as the probability of a long contiguous match across all decreases. For manual computation on small sets like the above, one approach is to generate all possible substrings from the shortest string (e.g., enumerate lengths from 1 upward in S3 = "BABA"), then check for exact consecutive presence in the others using string search (e.g., verify if "BA" appears contiguously in S1 and S2). This brute-force method has exponential time in string length but verifies the result transparently for illustration. The following table compares pairwise longest common substring lengths against the overall for the DNA-analogous example:| String Pair | LCS Length |
|---|---|
| S1 and S2 | 4 |
| S1 and S3 | 3 |
| S2 and S3 | 3 |
| All three (S1, S2, S3) | 3 |
Algorithms
Dynamic Programming Algorithm
The dynamic programming approach for finding the longest common substring (LCS) between two strings and of lengths and respectively utilizes a two-dimensional table to compute the lengths of common substrings ending at each pair of positions. The table represents the length of the longest common substring ending at position in and position in , where indices are 1-based for clarity.[9] The recurrence relation for filling the table is defined as follows: if , then ; otherwise, . This ensures that only contiguous matches are extended, distinguishing it from the longest common subsequence problem. The base cases are dp{{grok:render&&&type=render_inline_citation&&&citation_id=0&&&citation_type=wikipedia}} = 0 for all and dp{{grok:render&&&type=render_inline_citation&&&citation_id=0&&&citation_type=wikipedia}} = 0 for all , as no substring can end before the start of either string. The maximum value in the table gives the length of the LCS.[9] To retrieve the actual substring, backtracking starts from the cell with the maximum value and traces diagonally where characters match until reaching a zero. The following pseudocode illustrates the table construction and backtracking process:function LCSLengthAndSubstring(S, T):
m = length(S)
n = length(T)
dp = array of size (m+1) x (n+1), initialized to 0
max_length = 0
end_i = 0
end_j = 0
for i from 1 to m:
for j from 1 to n:
if S[i-1] == T[j-1]:
dp[i][j] = dp[i-1][j-1] + 1
if dp[i][j] > max_length:
max_length = dp[i][j]
end_i = i
end_j = j
else:
dp[i][j] = 0
// Backtrack to extract [substring](/page/Substring) from S
[substring](/page/Substring) = [empty string](/page/Empty_string)
i = end_i
j = end_j
while i > 0 and j > 0 and dp[i][j] > 0:
[substring](/page/Substring) = S[i-1] + [substring](/page/Substring)
i -= 1
j -= 1
return max_length, [substring](/page/Substring)
function LCSLengthAndSubstring(S, T):
m = length(S)
n = length(T)
dp = array of size (m+1) x (n+1), initialized to 0
max_length = 0
end_i = 0
end_j = 0
for i from 1 to m:
for j from 1 to n:
if S[i-1] == T[j-1]:
dp[i][j] = dp[i-1][j-1] + 1
if dp[i][j] > max_length:
max_length = dp[i][j]
end_i = i
end_j = j
else:
dp[i][j] = 0
// Backtrack to extract [substring](/page/Substring) from S
[substring](/page/Substring) = [empty string](/page/Empty_string)
i = end_i
j = end_j
while i > 0 and j > 0 and dp[i][j] > 0:
[substring](/page/Substring) = S[i-1] + [substring](/page/Substring)
i -= 1
j -= 1
return max_length, [substring](/page/Substring)
Suffix Tree Algorithm
The suffix tree algorithm for finding the longest common substring (LCS) between two strings and relies on constructing a generalized suffix tree for the concatenated string S\#T\$$, where #$STSTS#T$$.[10] The generalized suffix tree is built in linear time, specifically where and , using Ukkonen's on-line construction algorithm, which incrementally adds characters while maintaining the tree structure.[11] Once constructed, the LCS is found by traversing the tree to identify the deepest internal node whose subtree contains leaves from both and . The string depth (length of the path label from the root to that node) gives the LCS length, and the path label itself is the substring; this traversal visits each node once in time.[10] To compute the deepest such node, annotate each node during a post-order traversal by checking the origins of its leaf descendants. A node qualifies if its subtree includes at least one leaf from and one from , and among all qualifying nodes, the one with maximum string depth is selected. The following pseudocode outlines this process:function FindLCS(Tree tree, string S, string T):
max_depth = 0
lcs_node = null
function annotate(node v):
if v is leaf:
if v.suffix starts in S:
v.has_S = true
v.has_T = false
else:
v.has_S = false
v.has_T = true
return
v.has_S = false
v.has_T = false
for child in v.children:
annotate(child)
if child.has_S:
v.has_S = true
if child.has_T:
v.has_T = true
if v.has_S and v.has_T:
global max_depth, lcs_node
current_depth = string_depth(v)
if current_depth > max_depth:
max_depth = current_depth
lcs_node = v
annotate(root of tree)
return path_label(lcs_node)
function FindLCS(Tree tree, string S, string T):
max_depth = 0
lcs_node = null
function annotate(node v):
if v is leaf:
if v.suffix starts in S:
v.has_S = true
v.has_T = false
else:
v.has_S = false
v.has_T = true
return
v.has_S = false
v.has_T = false
for child in v.children:
annotate(child)
if child.has_S:
v.has_S = true
if child.has_T:
v.has_T = true
if v.has_S and v.has_T:
global max_depth, lcs_node
current_depth = string_depth(v)
if current_depth > max_depth:
max_depth = current_depth
lcs_node = v
annotate(root of tree)
return path_label(lcs_node)
Suffix Array Algorithm
The suffix array algorithm provides an efficient method for computing the longest common substring (LCS) between two or more strings by leveraging a sorted array of all suffixes from a concatenated input, avoiding the hierarchical structure of suffix trees. To apply this approach, the input strings are first concatenated into a single string of length (for two strings of lengths and ), separated by a unique sentinel character not present in the alphabet to distinguish suffixes from different original strings. The suffix array is then constructed by sorting all suffixes of in lexicographical order, which can be achieved in time using a comparison-based sorting algorithm that compares suffixes efficiently.[12][13] Once the suffix array is built, the longest common prefix (LCP) array is computed, where stores the length of the longest common prefix between the suffixes at positions and in the sorted order. This LCP array enables the identification of the LCS without explicit suffix comparisons. The LCP array can be constructed in linear time using Kasai's algorithm, which iterates through the suffixes in original order while using the suffix array to track previous matches and extend prefixes incrementally.[14] To find the LCS, the algorithm scans the LCP array for the maximum value such that the suffixes and originate from different original strings (i.e., they straddle the sentinel separator). The position of this maximum LCP indicates the starting index of the LCS in the concatenated string, and the value gives its length. This scanning step takes time, making the overall LCS computation dominated by the suffix array construction.[15] Kasai's algorithm for LCP computation proceeds as follows (pseudocode adapted for clarity):function KasaiLCP(S, SA, n):
rank = array of size n # inverse of SA
for i = 0 to n-1:
rank[SA[i]] = i
lcp = array of size n, initialized to 0
h = 0
for i = 0 to n-1:
if rank[i] > 0:
j = SA[rank[i] - 1]
while i + h < n and j + h < n and S[i + h] == S[j + h]:
h += 1
lcp[rank[i]] = h
if h > 0:
h -= 1
return lcp
function KasaiLCP(S, SA, n):
rank = array of size n # inverse of SA
for i = 0 to n-1:
rank[SA[i]] = i
lcp = array of size n, initialized to 0
h = 0
for i = 0 to n-1:
if rank[i] > 0:
j = SA[rank[i] - 1]
while i + h < n and j + h < n and S[i + h] == S[j + h]:
h += 1
lcp[rank[i]] = h
if h > 0:
h -= 1
return lcp






















