Recent from talks
Contribute something
Nothing was collected or created yet.
Blossom algorithm
View on WikipediaIn graph theory, the blossom algorithm is an algorithm for constructing maximum matchings on graphs. The algorithm was developed by Jack Edmonds in 1961,[1] and published in 1965.[2] Given a general graph G = (V, E), the algorithm finds a matching M such that each vertex in V is incident with at most one edge in M and |M| is maximized. The matching is constructed by iteratively improving an initial empty matching along augmenting paths in the graph. Unlike bipartite matching, the key new idea is that an odd-length cycle in the graph (blossom) is contracted to a single vertex, with the search continuing iteratively in the contracted graph.
The algorithm runs in time O(|E||V|2), where |E| is the number of edges of the graph and |V| is its number of vertices. A better running time of for the same task can be achieved with the much more complex algorithm of Micali and Vazirani.[3]
A major reason that the blossom algorithm is important is that it gave the first proof that a maximum-size matching could be found using a polynomial amount of computation time. Another reason is that it led to a linear programming polyhedral description of the matching polytope, yielding an algorithm for min-weight matching.[4] As elaborated by Alexander Schrijver, further significance of the result comes from the fact that this was the first polytope whose proof of integrality "does not simply follow just from total unimodularity, and its description was a breakthrough in polyhedral combinatorics."[5]
Augmenting paths
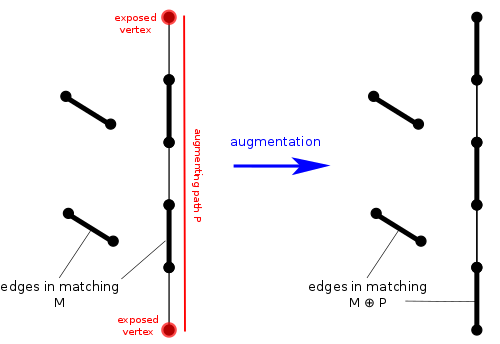
[edit]Given G = (V, E) and a matching M of G, a vertex v is exposed if no edge of M is incident with v. A path in G is an alternating path, if its edges are alternately not in M and in M (or in M and not in M). An augmenting path P is an alternating path that starts and ends at two distinct exposed vertices. Note that the number of unmatched edges in an augmenting path is greater by one than the number of matched edges, and hence the total number of edges in an augmenting path is odd. A matching augmentation along an augmenting path P is the operation of replacing M with a new matching
- .
By Berge's lemma, matching M is maximum if and only if there is no M-augmenting path in G.[6][7] Hence, either a matching is maximum, or it can be augmented. Thus, starting from an initial matching, we can compute a maximum matching by augmenting the current matching with augmenting paths as long as we can find them, and return whenever no augmenting paths are left. We can formalize the algorithm as follows:
INPUT: Graph G, initial matching M on G OUTPUT: maximum matching M* on G A1 function find_maximum_matching(G, M) : M* A2 P ← find_augmenting_path(G, M) A3 if P is non-empty then A4 return find_maximum_matching(G, augment M along P) A5 else A6 return M A7 end if A8 end function
We still have to describe how augmenting paths can be found efficiently. The subroutine to find them uses blossoms and contractions.
Blossoms and contractions
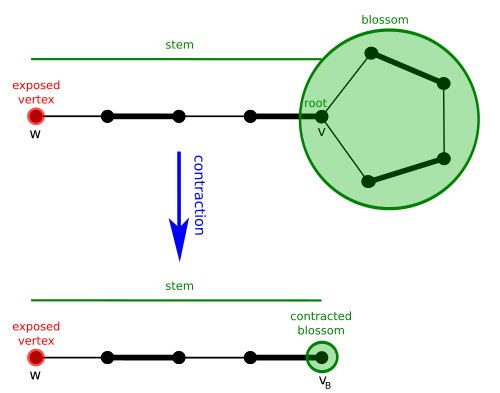
[edit]Given G = (V, E) and a matching M of G, a blossom B is a cycle in G consisting of 2k + 1 edges of which exactly k belong to M, and where one of the vertices v of the cycle (the base) is such that there exists an alternating path of even length (the stem) from v to an exposed vertex w.
Finding Blossoms:
- Traverse the graph starting from an exposed vertex.
- Starting from that vertex, label it as an outer vertex o.
- Alternate the labeling between vertices being inner i and outer o such that no two adjacent vertices have the same label.
- If we end up with two adjacent vertices labeled as outer o then we have an odd-length cycle and hence a blossom.
Define the contracted graph G' as the graph obtained from G by contracting every edge of B, and define the contracted matching M' as the matching of G' corresponding to M.
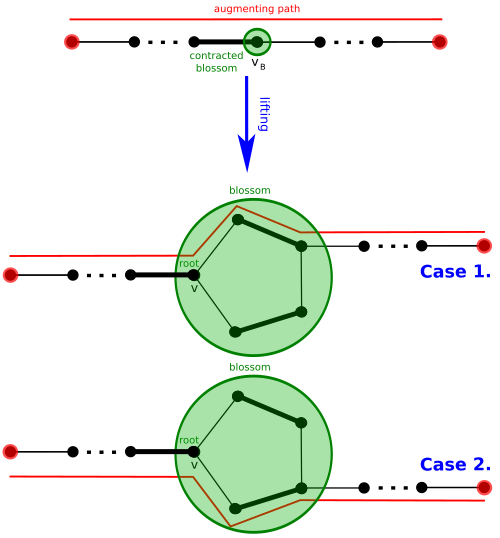
G' has an M'-augmenting path if and only if G has an M-augmenting path, and that any M'-augmenting path P' in G' can be lifted to an M-augmenting path in G by undoing the contraction by B so that the segment of P' (if any) traversing through vB is replaced by an appropriate segment traversing through B.[8] In more detail:
- if P' traverses through a segment u → vB → w in G', then this segment is replaced with the segment u → ( u' → … → w' ) → w in G, where blossom vertices u' and w' and the side of B, ( u' → … → w' ), going from u' to w' are chosen to ensure that the new path is still alternating (u' is exposed with respect to , ).
- if P' has an endpoint vB, then the path segment u → vB in G' is replaced with the segment u → ( u' → … → v' ) in G, where blossom vertices u' and v' and the side of B, ( u' → … → v' ), going from u' to v' are chosen to ensure that the path is alternating (v' is exposed, ).
Thus blossoms can be contracted and search performed in the contracted graphs. This reduction is at the heart of Edmonds' algorithm.
Finding an augmenting path
[edit]The search for an augmenting path uses an auxiliary data structure consisting of a forest F whose individual trees correspond to specific portions of the graph G. In fact, the forest F is the same that would be used to find maximum matchings in bipartite graphs (without need for shrinking blossoms). In each iteration the algorithm either (1) finds an augmenting path, (2) finds a blossom and recurses onto the corresponding contracted graph, or (3) concludes there are no augmenting paths. The auxiliary structure is built by an incremental procedure discussed next.[8]
The construction procedure considers vertices v and edges e in G and incrementally updates F as appropriate. If v is in a tree T of the forest, we let root(v) denote the root of T. If both u and v are in the same tree T in F, we let distance(u,v) denote the length of the unique path from u to v in T.
INPUT: Graph G, matching M on G
OUTPUT: augmenting path P in G or empty path if none found
B01 function find_augmenting_path(G, M) : P
B02 F ← empty forest
B03 unmark all vertices and edges in G, mark all edges of M
B05 for each exposed vertex v do
B06 create a singleton tree { v } and add the tree to F
B07 end for
B08 while there is an unmarked vertex v in F with distance(v, root(v)) even do
B09 while there exists an unmarked edge e = { v, w } do
B10 if w is not in F then
// w is matched, so add e and w's matched edge to F
B11 x ← vertex matched to w in M
B12 add edges { v, w } and { w, x } to the tree of v
B13 else
B14 if distance(w, root(w)) is odd then
// Do nothing.
B15 else
B16 if root(v) ≠ root(w) then
// Report an augmenting path in F { e }.
B17 P ← path (root(v) → ... → v) → (w → ... → root(w))
B18 return P
B19 else
// Contract a blossom in G and look for the path in the contracted graph.
B20 B ← blossom formed by e and edges on the path v → w in T
B21 G’, M’ ← contract G and M by B
B22 P’ ← find_augmenting_path(G’, M’)
B23 P ← lift P’ to G
B24 return P
B25 end if
B26 end if
B27 end if
B28 mark edge e
B29 end while
B30 mark vertex v
B31 end while
B32 return empty path
B33 end function
Examples
[edit]The following four figures illustrate the execution of the algorithm. Dashed lines indicate edges that are currently not present in the forest. First, the algorithm processes an out-of-forest edge that causes the expansion of the current forest (lines B10 – B12).
Next, it detects a blossom and contracts the graph (lines B20 – B21).
Finally, it locates an augmenting path P′ in the contracted graph (line B22) and lifts it to the original graph (line B23). Note that the ability of the algorithm to contract blossoms is crucial here; the algorithm cannot find P in the original graph directly because only out-of-forest edges between vertices at even distances from the roots are considered on line B17 of the algorithm.
Analysis
[edit]The forest F constructed by the find_augmenting_path() function is an alternating forest.[9]
- a tree T in G is an alternating tree with respect to M, if
- T contains exactly one exposed vertex r called the tree root
- every vertex at an odd distance from the root has exactly two incident edges in T, and
- all paths from r to leaves in T have even lengths, their odd edges are not in M and their even edges are in M.
- a forest F in G is an alternating forest with respect to M, if
- its connected components are alternating trees, and
- every exposed vertex in G is a root of an alternating tree in F.
Each iteration of the loop starting at line B09 either adds to a tree T in F (line B10) or finds an augmenting path (line B17) or finds a blossom (line B20). It is easy to see that the running time is .
Bipartite matching
[edit]When G is bipartite, there are no odd cycles in G. In that case, blossoms will never be found and one can simply remove lines B20 – B24 of the algorithm. The algorithm thus reduces to the standard algorithm to construct maximum cardinality matchings in bipartite graphs[7] where we repeatedly search for an augmenting path by a simple graph traversal: this is for instance the case of the Ford–Fulkerson algorithm.
Weighted matching
[edit]The matching problem can be generalized by assigning weights to edges in G and asking for a set M that produces a matching of maximum (minimum) total weight: this is the maximum weight matching problem. This problem can be solved by a combinatorial algorithm that uses the unweighted Edmonds's algorithm as a subroutine.[6] Vladimir Kolmogorov provides an efficient C++ implementation of this.[10]
References
[edit]- ^ Edmonds, Jack (1991), "A glimpse of heaven", in J.K. Lenstra; A.H.G. Rinnooy Kan; A. Schrijver (eds.), History of Mathematical Programming --- A Collection of Personal Reminiscences, CWI, Amsterdam and North-Holland, Amsterdam, pp. 32–54
- ^ Edmonds, Jack (1965). "Paths, trees, and flowers". Can. J. Math. 17: 449–467. doi:10.4153/CJM-1965-045-4.
- ^ Micali, Silvio; Vazirani, Vijay (1980). An O(V1/2E) algorithm for finding maximum matching in general graphs. 21st Annual Symposium on Foundations of Computer Science. IEEE Computer Society Press, New York. pp. 17–27.
- ^ Edmonds, Jack (1965). "Maximum matching and a polyhedron with 0,1-vertices". Journal of Research of the National Bureau of Standards Section B. 69: 125–130. doi:10.6028/jres.069B.013.
- ^ Schrijver, Alexander (2003). Combinatorial Optimization: Polyhedra and Efficiency. Algorithms and Combinatorics. Berlin Heidelberg: Springer-Verlag. ISBN 9783540443896.
- ^ a b Lovász, László; Plummer, Michael (1986). Matching Theory. Akadémiai Kiadó. ISBN 963-05-4168-8.
- ^ a b Karp, Richard, "Edmonds's Non-Bipartite Matching Algorithm", Course Notes. U. C. Berkeley (PDF), archived from the original (PDF) on 2008-12-30
- ^ a b Tarjan, Robert, "Sketchy Notes on Edmonds' Incredible Shrinking Blossom Algorithm for General Matching", Course Notes, Department of Computer Science, Princeton University (PDF)
- ^ Kenyon, Claire; Lovász, László, "Algorithmic Discrete Mathematics", Technical Report CS-TR-251-90, Department of Computer Science, Princeton University
- ^ Kolmogorov, Vladimir (2009), "Blossom V: A new implementation of a minimum cost perfect matching algorithm", Mathematical Programming Computation, 1 (1): 43–67, doi:10.1007/s12532-009-0002-8
Blossom algorithm
View on GrokipediaBackground
Graph matching basics
In graph theory, an undirected graph consists of a vertex set and an edge set , where edges connect pairs of vertices. A matching in is a subset of edges such that no two edges in share a common vertex.[4][5] A maximum matching in a graph is a matching that contains the largest possible number of edges among all matchings in that graph.[6][7] A perfect matching is a special case of a maximum matching where every vertex in is incident to exactly one edge in the matching, which requires to be even.[8][9] The maximum matching problem seeks to find a maximum matching in an undirected graph , a fundamental combinatorial optimization task with applications in resource allocation and network design.[10] Early foundational work on this problem for bipartite graphs was established by Dénes Kőnig in 1916, who proved that the size of a maximum matching equals the minimum vertex cover in such graphs.[11][12]Bipartite versus general matching
A bipartite graph is an undirected graph whose vertices can be divided into two disjoint sets such that no two graph vertices in the same set are adjacent.[13] In such graphs, the size of a maximum matching equals the size of a minimum vertex cover, as established by König's theorem.[13] This duality enables efficient algorithms for computing maximum matchings in bipartite graphs, including the Hungarian algorithm, which runs in O(n^3) time for n vertices,[14] and the Hopcroft-Karp algorithm, which achieves O(\sqrt{n} m) time for m edges.[15] In general graphs, however, König's theorem does not hold, and the size of a maximum matching can be strictly smaller than the size of a minimum vertex cover—for instance, in a triangle graph, the maximum matching has size 1 while the minimum vertex cover has size 2.[16] Moreover, the presence of odd cycles in general graphs complicates the search for augmenting paths, as these structures can trap the search process without revealing all possible improvements to the matching.[16] Berge's lemma provides a foundational characterization for general graphs: a matching is maximum if and only if no augmenting path exists with respect to that matching.[17] This result underscores the need for algorithms that systematically handle cyclic structures in non-bipartite graphs to ensure completeness in finding augmenting paths.Core Concepts
Augmenting paths
In graph theory, a matching is a set of edges without common vertices. An augmenting path relative to a matching in an undirected graph is a simple path that starts and ends at unmatched vertices (also called free or exposed vertices) and whose edges alternate between those not in and those in ; specifically, it begins and ends with an edge not in .[18][19] The presence of an augmenting path allows the matching to be extended by taking the symmetric difference , which consists of all edges in or but not both. This operation flips the status of edges along : unmatched edges become matched, and matched edges become unmatched, resulting in a new matching whose size is exactly one greater than , since contributes one more unmatched edge than matched edges.[20] Formally, for any augmenting path relative to .[21] To illustrate, consider a path where and are unmatched, is matched in , and , are unmatched; flipping yields a matching with edges and , augmenting the size by one. In bipartite graphs, augmenting paths can be efficiently located using breadth-first search (BFS) or depth-first search (DFS) from free vertices in one part, following alternating edges without complications from cycles.[22] This process repeatedly augments the matching until no such paths remain, yielding a maximum matching. The Tutte–Berge formula provides a global characterization of the maximum matching size in general graphs: for a graph with , the size of a maximum matching is , where denotes the number of odd-sized connected components in the subgraph induced by .[23] This formula, established in 1958, equates the maximum matching size to half the minimum deficiency over all vertex subsets .[24]Blossoms and odd cycles
In the context of maximum matching in general graphs, a blossom is defined as an odd-length alternating cycle with respect to a given matching , consisting of vertices for some integer , where exactly edges belong to , leaving one vertex unmatched within the cycle, known as the base or stem vertex.[25] This structure arises specifically in non-bipartite graphs, where the presence of odd cycles distinguishes the problem from bipartite matching.[26] Odd cycles pose a significant challenge to the standard augmenting path method because they permit multiple alternating paths from a free vertex to converge on the same vertex, thereby violating the assumption of simple paths and potentially leading to incorrect or incomplete searches for augmentations.[27] In particular, traversing an odd cycle in either direction can create the illusion of an augmenting path that does not truly increase the matching size, as the cycle's odd length ensures an imbalance in matched and unmatched edges incident to the base vertex.[24] A key property of blossoms is their amenability to contraction, wherein the entire odd cycle is collapsed into a single supernode, preserving the equivalence of maximum matchings between the original graph and the contracted one.[25] This contraction simplifies the graph structure without losing information about potential augmenting paths external to the blossom. In his seminal 1965 work, Jack Edmonds observed that to correctly identify true augmenting paths in general graphs, such blossoms must be systematically detected and shrunk, enabling the algorithm to proceed as if in a bipartite setting after handling these obstacles.[28] For illustration, consider a 5-cycle (a blossom with ) where vertices are labeled in sequence, with matched edges and in , leaving as the free stem vertex connected by unmatched edges to and .[27] This configuration blocks straightforward path extension until contraction occurs. The concept of blossoms also connects to broader matching theory, as W. T. Tutte's 1947 theorem characterizes the existence of perfect matchings in general graphs via a condition on odd components in barrier sets, with blossoms representing local manifestations of such structural barriers that Edmonds' approach resolves algorithmically.[29]Algorithm Mechanics
Overview of the procedure
The Blossom algorithm, developed by Jack Edmonds, aims to compute a maximum cardinality matching in non-bipartite undirected graphs, where a matching is a set of edges without common vertices, and maximum cardinality means the largest possible number of such edges.[2] The input is an undirected graph , with as the vertex set and as the edge set, and the output is a maximum matching .[2] Published in 1965, this algorithm marked the first polynomial-time solution for the general matching problem, extending beyond bipartite cases that were solvable earlier via simpler methods.[2] At a high level, the procedure operates iteratively: starting from an initial (possibly empty) matching , it repeatedly searches for an augmenting path relative to —a path that alternates between edges not in and edges in , connecting two unmatched vertices.[2] If such a path is found, the matching is augmented by flipping the edges along it (adding the non-matching edges and removing the matching ones), increasing the matching size by one.[2] In non-bipartite graphs, searches may be blocked by odd cycles known as blossoms; the algorithm addresses this by contracting these blossoms into single vertices, effectively recursing on a modified graph to continue the search.[2] This process repeats until no augmenting path exists, at which point is maximum.[2] A skeletal pseudocode for the algorithm captures this iterative structure:function BlossomAlgorithm(G = (V, E)):
M ← ∅ // Initial empty matching
while there exists an augmenting path P relative to M:
augment M along P // Flip edges in P to update M
if blossom encountered during search:
contract blossom and recurse on reduced graph
return M
function BlossomAlgorithm(G = (V, E)):
M ← ∅ // Initial empty matching
while there exists an augmenting path P relative to M:
augment M along P // Flip edges in P to update M
if blossom encountered during search:
contract blossom and recurse on reduced graph
return M
Blossom detection and contraction
In the Blossom algorithm, blossoms are detected during the search for augmenting paths, which proceeds from free (unmatched) vertices using a depth-first or breadth-first search to build alternating paths in the graph relative to the current matching. Specifically, a blossom—an odd-length alternating cycle—is identified when a back edge connects two vertices at even distances from the root in the search tree, indicating the presence of an odd cycle that obstructs the bipartite assumption.[26][27] To manage the search structure, vertices are labeled as outer or inner based on their role in the alternating paths and contractions. Outer vertices include those that are unmatched or reached through previously identified augmenting paths, forming the "exposed" layer for continued search; inner vertices, conversely, lie within contracted blossoms and are treated as part of the supervertex to avoid revisiting the cycle's internal structure. This labeling ensures that the search alternates between matched and unmatched edges while respecting the contracted components.[26][30] Upon detection, the blossom is contracted by shrinking the entire odd cycle into a single supervertex in a new auxiliary graph , where the vertices of the cycle are merged while preserving the internal matching edges within the supervertex. Edges incident to the cycle are adjusted to connect to this supervertex: an edge between two cycle vertices becomes internal (and thus ignored for external search), while edges from outside the cycle attach to the supervertex. The contracted graph has a reduced matching that excludes the blossom's internal edges but maintains the overall matching size.[26][27] This contraction preserves equivalence for augmenting path searches because any augmenting path in the original graph corresponds to one in , and vice versa; upon finding a path in , it can be "lifted" or expanded back into by traversing the blossom's alternating cycle appropriately, ensuring the path remains augmenting. The process terminates when no further augmenting paths exist, with each contraction reducing the graph size by at least two vertices.[26][2] Multiple blossoms are handled through nested and layered contractions, where newly detected blossoms may form within or around previously contracted ones, treated as a hierarchy of supervertices during recursive searches. Each augmentation phase may involve up to contractions, but the overall algorithm bounds the total by polynomial time through careful expansion during path lifting.[26][30]Searching for augmenting paths
The search for augmenting paths in the Blossom algorithm begins with an exposed (unmatched) vertex, which serves as the root of an alternating forest. A depth-first search (DFS) is employed to explore alternating paths, where vertices are labeled with even or odd parity based on their distance from the root, ensuring the path alternates between matched and unmatched edges.[26][31] This labeling adapts the bipartite case by building a forest rather than a single tree, allowing multiple exposed vertices to be roots and preventing cycles within individual trees.[32] During the DFS traversal, unmarked edges incident to labeled vertices are examined. If an edge connects to an unlabeled exposed vertex, an augmenting path is immediately found. Similarly, connecting to a vertex labeled even with a different root yields an augmenting path by combining the two even-length alternating paths from their roots. However, if the edge connects two vertices with the same root and even labels, it indicates an odd-length alternating cycle, known as a blossom, which must be handled to continue the search.[33][26] The search integrates blossom detection by identifying the cycle formed by the path from the lowest common ancestor to the two even-labeled vertices and the connecting edge.[32] Upon detecting a blossom, the algorithm contracts it into a single supernode, typically the base vertex (the odd-labeled vertex closest to the root in the cycle), reducing the graph while preserving the existence of augmenting paths. The search then recurses on this contracted graph, continuing the DFS from the appropriate point. This contraction temporarily simplifies the structure, allowing the exploration of alternating paths in the reduced graph without losing potential augmentations.[33][31] Key data structures support this process, including adjacency lists for efficient edge traversal, a matching array to track paired vertices, and blossom representatives to record contractions and maintain the hierarchy of supernodes.[26] If an augmenting path is found in the contracted graph, an expansion phase lifts it back to the original graph. This involves traversing the contraction history in reverse order, selecting the appropriate base for each blossom—specifically, the one that connects to the path via an unmatched edge—and inserting the even-length alternating path within the blossom to preserve the augmenting property. The resulting path in the original graph is then used to augment the matching by flipping matched and unmatched edges along it, increasing the matching size by one.[33][26] The search procedure repeats from newly exposed vertices until no further augmenting path can be found, at which point the current matching is maximum, as per Berge's lemma generalized to non-bipartite graphs. Termination occurs when all exposed vertices have been processed without discovering a path or when the forest covers all possible alternating structures without blossoms yielding augmentations.[2][32]Examples and Illustrations
Bipartite matching walkthrough
Consider a simple bipartite graph with partite sets and , connected by the edges -, -, -, -, -, and -.[34] The Blossom algorithm starts with an empty matching of size 0. In the first iteration, perform a BFS from the free vertices in to find an augmenting path. An augmenting path is -, which alternates between unmatched and (trivially) matched edges. Augment the matching by including this edge, resulting in a matching of size 1: -.[28] In the second iteration, BFS from the remaining free vertices in identifies the path -. Augment by adding this edge, yielding a matching of size 2: --. The current matching size has increased, and the previously free is now matched.[28] In the third iteration, BFS uncovers the path -. Augmenting along this path produces a matching of size 3: ---. No further augmenting paths exist, as subsequent BFS searches from any remaining free vertices (none in ) terminate without reaching a free vertex in .[28] This example demonstrates the before-and-after progression: the initial empty matching grows incrementally via each augmenting path, with the final matching covering all vertices in but leaving unmatched. In bipartite graphs, the Blossom algorithm encounters no blossoms, as the absence of odd cycles eliminates the need for detection and contraction; it reduces to standard augmenting path searches.[26] This aligns with simpler bipartite methods like the Hungarian algorithm.[35] The result is a maximum matching of size 3 without any contractions. In the bipartite case, the algorithm achieves this in time, with at most augmentations and each BFS taking time.[35]Non-bipartite example with blossom
Consider a representative non-bipartite graph with vertices and edges , , , , . The initial matching is , leaving , , and exposed.[36] This setup features an odd cycle --- that forms a blossom during the search for an augmenting path. The algorithm begins the search for an augmenting path from the exposed vertex using a layered alternating tree. Place at layer 0 (even). From layer 0, traverse the unmatched edge - to reach at layer 1 (odd). From layer 1, traverse the matched edge - to reach at layer 2 (even). From layer 2, traverse the unmatched edge - to reach at layer 3 (odd). At this point, from (layer 3, odd), there is an unmatched edge -, where is already in the tree at layer 1 (odd). Since both endpoints have odd parity, this detects a blossom: the odd cycle --- with base vertex (the lowest-layer vertex in the cycle).[36] To resolve the blossom, contract the cycle -- into a single supervertex . The contracted graph now has vertices , , and edges - (from original -), - (from original -). The internal matching edge - is preserved within , leaving exposed in the contracted graph (as no external matched edges incident to the blossom vertices). Continue the search in this reduced graph from at layer 0. Traverse the unmatched edge - to reach at layer 1 (odd). Since is exposed and reached at an odd layer, an augmenting path - is found. The original graph before contraction is shown below in simple ASCII representation: s --- a --- b
| /
c /
|
t
Edges: s-a, a-b, b-c, c-a, c-t
Matched: a-b (solid), others unmatched (dashed implied).
s --- a --- b
| /
c /
|
t
Edges: s-a, a-b, b-c, c-a, c-t
Matched: a-b (solid), others unmatched (dashed implied).
s --- S --- t
Supervertex S = {a,b,c}
s --- S --- t
Supervertex S = {a,b,c}
s - a
\
b - c - t
Matched edges: s-a, b-c (solid).
s - a
\
b - c - t
Matched edges: s-a, b-c (solid).
Analysis and Properties
Time complexity
The original Blossom algorithm, as proposed by Edmonds, exhibits a time complexity of , stemming from up to augmenting path searches, each taking time due to the naive handling of blossom contractions and path explorations in dense graphs.[37] Subsequent implementations refined this to or by optimizing data structures for labeling and searching, but the fundamental bound remains quartic in the worst case for the basic procedure.[38] Significant asymptotic improvements came with the Micali-Vazirani algorithm in 1980, achieving time by generalizing Hopcroft-Karp-style phase-based augmentations to handle multiple shortest augmenting paths simultaneously while efficiently managing blossoms through layered searches.[39] This bound, equivalent to , represents the best known theoretical complexity for maximum cardinality matching in general graphs and incorporates ideas from bipartite matching to reduce the number of phases to . The breakdown highlights the searching for augmenting paths as the primary bottleneck, where each phase processes edges in phases. Blossom detection and contraction operations are optimized using union-find-like data structures with path compression and union-by-rank, enabling near-constant time merges and finds for maintaining contracted components during searches.[40] The space complexity is , sufficient for adjacency lists, matching labels, and auxiliary trees without excessive overhead.[3] Compared to the time of the Hopcroft-Karp algorithm for bipartite matching, the general Blossom variants are asymptotically slower due to the added complexity of odd cycles, yet they provide the first polynomial-time solution for non-bipartite graphs. No major asymptotic advancements have emerged since the 1980s, though practical implementations incorporate heuristics and refined data structures for better real-world performance on sparse graphs.[3]Correctness proof outline
The correctness of the Blossom algorithm, also known as Edmonds' matching algorithm, relies fundamentally on Berge's lemma, which states that a matching in a graph is maximum if and only if there exists no augmenting path relative to that matching. The algorithm iteratively searches for augmenting paths and augments the matching upon finding one, halting when no such path exists in the current graph, thereby ensuring the final matching is maximum by Berge's lemma.[41] A key invariant in the proof is that contractions of blossoms preserve the existence of augmenting paths: if an augmenting path exists in the original graph, a corresponding augmenting path exists in the contracted graph, and vice versa.[36] This preservation holds because shrinking an odd-length alternating cycle (blossom) into a single vertex maintains the parity and connectivity properties necessary for path existence, allowing the search to proceed equivalently in the reduced graph.[42] Blossom handling ensures no false augmenting paths are created during shrinking, as any path found in the contracted graph can be expanded back to a valid alternating path in the original graph without introducing cycles that violate matching properties.[41] Upon detection of a blossom's tip, the algorithm contracts it, and when an augmenting path reaches the contracted vertex, expansion along the blossom's alternating cycle recovers the true augmenting path in the original structure.[36] The proof proceeds by induction on the number of augmentations: each successful augmentation strictly increases the matching size by one, and the process terminates with no augmenting path, invoking Berge's lemma to confirm maximality. Since the algorithm starts from an empty or initial matching and augments until impossible, the final matching is maximum by the inductive application of Berge's characterization.[41] The algorithm implicitly verifies the conditions of Tutte's theorem, which characterizes the existence of perfect matchings via the absence of Tutte sets (subsets where the number of odd components exceeds the set's size), as the failure to find an augmenting path corresponds to a barrier structure aligning with the Tutte-Berge formula for maximum matching size.[36] Specifically, the proof in Edmonds' original work demonstrates that paths in the contracted graph lift correctly to the original, ensuring the algorithm's outputs satisfy the structural conditions for optimality.Extensions and Applications
Weighted matching variants
The weighted maximum matching problem extends the unweighted Blossom algorithm to graphs with edge weights, seeking a matching that maximizes the sum of the weights of its edges.[43] This formulation applies to real-valued weights and is central to optimization tasks where edge costs represent profits, similarities, or other quantitative measures.[43] Edmonds developed the weighted variant of the Blossom algorithm in 1965, building on his earlier work for cardinality matching.[43] The algorithm employs a primal-dual approach, introducing dual variables—node potentials for vertices and set variables for blossoms —to track optimality conditions. These potentials adjust reduced edge costs , ensuring non-negative costs in an auxiliary graph while maintaining feasibility.[3] A key theoretical foundation is the linear programming relaxation of the matching polyhedron, strengthened by blossom inequalities: for any odd-sized subset of vertices with , the inequality ensures integrality, where are edge variables. The algorithmic structure mirrors the unweighted Blossom procedure but incorporates weight optimization through repeated shortest-path computations in the auxiliary graph.[3] Augmenting paths are found using Dijkstra's algorithm with potentials to efficiently handle reduced costs, followed by blossom detection and contraction as in the cardinality case.[3] Dual variables are updated after each augmentation to restore optimality, with the process repeating until no augmenting path exists.[3] This yields a maximum-weight matching while satisfying the blossom inequalities at equality for the optimal solution. Unlike the unweighted algorithm, which focuses solely on cardinality, the weighted version accommodates negative edge weights through reductions to non-negative cases or direct handling in the primal-dual framework, enabling broader applications such as the assignment problem in bipartite graphs.[3] In the bipartite special case, it reduces to the Hungarian algorithm, solving the linear assignment problem efficiently. Early implementations run in O(|V|^4) time, but subsequent optimizations achieve O(|V|^3) time, where |V| is the number of vertices, with modern variants like Blossom V achieving similar or improved practical performance through optimized data structures such as Fibonacci heaps for Dijkstra searches.[3]Practical implementations
The Blossom algorithm has been implemented in several open-source libraries, enabling its use in practical computing environments. The Lemon graph library, written in C++, incorporates the Blossom V implementation, which computes maximum weight matchings in general graphs using priority queues and variable dual updates for efficiency.[3] Similarly, the Python library NetworkX provides a Blossom-based solver via itsmax_weight_matching function, supporting unweighted and weighted cases for general graphs and integrating seamlessly with other graph analysis tools. Open-source implementations of the algorithm, including early variants, have been publicly available since the 1990s through research repositories and graph software distributions.
Optimizations in these implementations often draw from Gabow's refinement, which achieves O(|V|^3) time complexity through structured handling of blossoms and efficient path searches, making it suitable for graphs up to thousands of vertices.[38] For small to medium graphs, adjacency matrix representations are commonly employed to simplify blossom contraction and adjacency queries, trading space for faster access in dense cases.[3]
In applications, the algorithm supports pairing in molecular chemistry via dimer models, where it enumerates perfect matchings to model molecular dimer configurations on lattice graphs.[44] It also aids computer vision tasks like stereo matching by computing correspondences in non-bipartite feature graphs.[45] In economics, extensions to the stable marriage problem on general graphs leverage Blossom for finding stable matchings beyond bipartite settings.[46] Additionally, bioinformatics uses it for RNA folding, where maximum matching predicts pseudoknot-free secondary structures by pairing complementary bases.[47] Recent adaptations like Sparse Blossom (2025) apply the algorithm to quantum error correction by computing minimum-weight perfect matchings for syndrome decoding in surface codes.[48]
Scalability poses challenges for large graphs, as the cubic complexity limits exact solutions to instances with up to 10^4 vertices on standard hardware, prompting approximations such as the greedy algorithm, which provides a 1/2-approximation, or (1-ε)-approximations running in near-linear time for graphs with millions of vertices.[49] Recent developments post-2020 include parallel implementations integrated into machine learning frameworks, such as X-Blossom for accelerating graph matching in graph neural networks and economic analysis pipelines.[45]