Recent from talks
All channels
Be the first to start a discussion here.
Be the first to start a discussion here.
Be the first to start a discussion here.
Be the first to start a discussion here.
Welcome to the community hub built to collect knowledge and have discussions related to Reporter gene.
Nothing was collected or created yet.
Reporter gene
View on Wikipediafrom Wikipedia
Not found
Reporter gene
View on Grokipediafrom Grokipedia
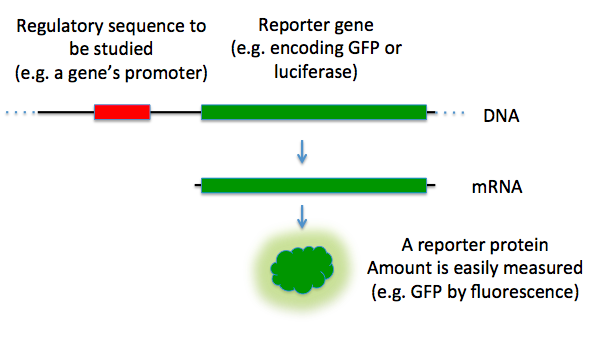
A reporter gene is a gene that encodes a protein whose expression can be readily and specifically detected, often through enzymatic activity, fluorescence, or luminescence, allowing scientists to monitor the activity of promoters, enhancers, or other regulatory elements in molecular biology experiments.[1] These genes are typically fused to sequences of interest or used as selectable markers to track gene expression patterns, cellular transformations, and biological processes in living systems.[2] By producing distinguishable signals from endogenous proteins, reporter genes provide a sensitive and quantifiable readout of transcriptional regulation, making them indispensable tools in biotechnology and genetic research.[3]
The concept of reporter genes emerged in the early 1980s with the adoption of the bacterial chloramphenicol acetyltransferase (CAT) gene to measure promoter activity in mammalian cells, marking a shift from indirect methods like Northern blotting to direct, enzyme-based assays.[4] This was followed by the widespread use of the Escherichia coli lacZ gene, encoding β-galactosidase, which produces a blue color upon substrate hydrolysis and has been a staple for visualizing gene expression in tissues and cells due to its stability and ease of detection.[1] The discovery and engineering of green fluorescent protein (GFP) from the jellyfish Aequorea victoria in the 1960s, with practical applications expanding in the 1990s, revolutionized non-invasive imaging by enabling real-time visualization of gene expression without substrates.[4] Other prominent examples include luciferase from fireflies or sea pansy, which catalyzes a bioluminescent reaction for high-sensitivity, low-background detection in live animals and high-throughput screens.[3]
Reporter genes find broad applications in studying gene regulation, such as in promoter-reporter constructs where expression levels correlate with transcriptional activation or repression in response to stimuli like hormones or drugs.[3] In molecular imaging, they facilitate non-invasive tracking of cell migration, tumor growth, and gene therapy efficacy using techniques like fluorescence microscopy, bioluminescence imaging, or positron emission tomography (PET) when paired with complementary probes.[4] For instance, GFP variants and luciferase have been integrated into transgenic mouse models to trace developmental lineages or immune responses, while β-galactosidase remains favored for histological analysis in fixed tissues.[1] These tools also support drug discovery by evaluating bioactivity in reporter gene assays, such as measuring pathway activation in cell lines for biologics like monoclonal antibodies.[3] Despite their utility, challenges include potential immunogenicity of foreign proteins and the need for optimized delivery vectors to ensure accurate, cell-specific expression.[4]