Recent from talks
Nothing was collected or created yet.
Inverted repeat
View on WikipediaAn inverted repeat (or IR) is a single stranded sequence of nucleotides followed downstream by its reverse complement.[1] The intervening sequence of nucleotides between the initial sequence and the reverse complement can be any length including zero. For example, 5'---TTACGnnnnnnCGTAA---3' is an inverted repeat sequence. When the intervening length is zero, the composite sequence is a palindromic sequence.[2]
Both inverted repeats and direct repeats constitute types of nucleotide sequences that occur repetitively. These repeated DNA sequences often range from a pair of nucleotides to a whole gene, while the proximity of the repeat sequences varies between widely dispersed and simple tandem arrays.[3] The short tandem repeat sequences may exist as just a few copies in a small region to thousands of copies dispersed all over the genome of most eukaryotes.[4] Repeat sequences with about 10–100 base pairs are known as minisatellites, while shorter repeat sequences having mostly 2–4 base pairs are known as microsatellites.[5] The most common repeats include the dinucleotide repeats, which have the bases AC on one DNA strand, and GT on the complementary strand.[3] Some elements of the genome with unique sequences function as exons, introns and regulatory DNA.[6] Though the most familiar loci of the repetitive sequences are the centromere and the telomere,[6] a large portion of the repeated sequences in the genome are found among the noncoding DNA.[5]
Inverted repeats have a number of important biological functions. They define the boundaries in transposons and indicate regions capable of self-complementary base pairing (regions within a single sequence which can base pair with each other). These properties play an important role in genome instability[7] and contribute not only to cellular evolution and genetic diversity[8] but also to mutation and disease.[9] In order to study these effects in detail, a number of programs and databases have been developed to assist in discovery and annotation of inverted repeats in various genomes.
Understanding inverted repeats
[edit]Example of an inverted repeat
[edit]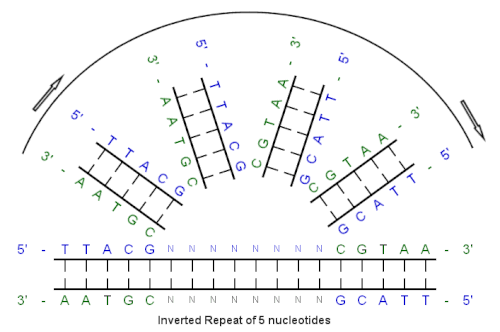
Beginning with this initial sequence:
5'-TTACG-3'
The complement created by base pairing is:
3'-AATGC-5'
The reverse complement is:
5'-CGTAA-3'
And, the inverted repeat sequence is:
5'---TTACGnnnnnnCGTAA---3'
"nnnnnn" represents any number of intervening nucleotides.
Vs. direct repeat
[edit]A direct repeat occurs when a sequence is repeated with the same pattern downstream.[1] There is no inversion and no reverse complement associated with a direct repeat. The nucleotide sequence written in bold characters signifies the repeated sequence. It may or may not have intervening nucleotides.
- 5´ TTACGnnnnnnTTACG 3´
- 3´ AATGCnnnnnnAATGC 5´
Linguistically, a typical direct repeat is comparable to rhyming, as in "time on a dime".
Vs. tandem repeat
[edit]A direct repeat with no intervening nucleotides between the initial sequence and its downstream copy is a Tandem repeat. The nucleotide sequence written in bold characters signifies the repeated sequence.
- 5´ TTACGTTACG 3´
- 3´ AATGCAATGC 5´
Linguistically, a typical tandem repeat is comparable to stuttering, or deliberately repeated words, as in "bye-bye".
Vs. palindrome
[edit]An inverted repeat sequence with no intervening nucleotides between the initial sequence and its downstream reverse complement is a palindrome.[1]
EXAMPLE:
Step 1: start with an inverted repeat: 5' TTACGnnnnnnCGTAA 3'
Step 2: remove intervening nucleotides: 5' TTACGCGTAA 3'
This resulting sequence is palindromic because it is the reverse complement of itself.[1]
- 5' TTACGCGTAA 3' test sequence (from Step 2 with intervening nucleotides removed)
- 3' AATGCGCATT 5' complement of test sequence
- 5' TTACGCGTAA 3' reverse complement This is the same as the test sequence above, and thus, it is a palindrome.
Biological features and functionality
[edit]Conditions that favor synthesis
[edit]The diverse genome-wide repeats are derived from transposable elements, which are now understood to "jump" about different genomic locations, without transferring their original copies.[10] Subsequent shuttling of the same sequences over numerous generations ensures their multiplicity throughout the genome.[10] The limited recombination of the sequences between two distinct sequence elements known as conservative site-specific recombination (CSSR) results in inversions of the DNA segment, based on the arrangement of the recombination recognition sequences on the donor DNA and recipient DNA.[10] Again, the orientation of two of the recombining sites within the donor DNA molecule relative to the asymmetry of the intervening DNA cleavage sequences, known as the crossover region, is pivotal to the formation of either inverted repeats or direct repeats.[10] Thus, recombination occurring at a pair of inverted sites will invert the DNA sequence between the two sites.[10] Very stable chromosomes have been observed with comparatively fewer numbers of inverted repeats than direct repeats, suggesting a relationship between chromosome stability and the number of repeats.[11]
Regions where presence is obligatory
[edit]Terminal inverted repeats have been observed in the DNA of various eukaryotic transposons, even though their source remains unknown.[12] Inverted repeats are principally found at the origins of replication of cell organism and organelles that range from phage plasmids, mitochondria, and eukaryotic viruses to mammalian cells.[13] The replication origins of the phage G4 and other related phages comprise a segment of nearly 139 nucleotide bases that include three inverted repeats that are essential for replication priming.[13]
In the genome
[edit]To a large extent, portions of nucleotide repeats are quite often observed as part of rare DNA combinations.[14] The three main repeats which are largely found in particular DNA constructs include the closely precise homopurine-homopyrimidine inverted repeats, which is otherwise referred to as H palindromes, a common occurrence in triple helical H conformations that may comprise either the TAT or CGC nucleotide triads. The others could be described as long inverted repeats having the tendency to produce hairpins and cruciform, and finally direct tandem repeats, which commonly exist in structures described as slipped-loop, cruciform and left-handed Z-DNA.[14]
Common in different organisms
[edit]Past studies suggest that repeats are a common feature of eukaryotes unlike the prokaryotes and archaea.[14] Other reports suggest that irrespective of the comparative shortage of repeat elements in prokaryotic genomes, they nevertheless contain hundreds or even thousands of large repeats.[15] Current genomic analysis seem to suggest the existence of a large excess of perfect inverted repeats in many prokaryotic genomes as compared to eukaryotic genomes.[16]

For quantification and comparison of inverted repeats between several species, namely on archaea, see [17]
Inverted repeats in pseudoknots
[edit]Pseudoknots are common structural motifs found in RNA. They are formed by two nested stem-loops such that the stem of one structure is formed from the loop of the other. There are multiple folding topologies among pseudoknots and great variation in loop lengths, making them a structurally diverse group.[18]
Inverted repeats are a key component of pseudoknots as can be seen in the illustration of a naturally occurring pseudoknot found in the human telomerase RNA component.[19] Four different sets of inverted repeats are involved in this structure. Sets 1 and 2 are the stem of stem-loop A and are part of the loop for stem-loop B. Similarly, sets 3 and 4 are the stem for stem-loop B and are part of the loop for stem-loop A.
Pseudoknots play a number of different roles in biology. The telomerase pseudoknot in the illustration is critical to that enzyme's activity.[19] The ribozyme for the hepatitis delta virus (HDV) folds into a double-pseudoknot structure and self-cleaves its circular genome to produce a single-genome-length RNA. Pseudoknots also play a role in programmed ribosomal frameshifting found in some viruses and required in the replication of retroviruses.[18]
In riboswitches
[edit]Inverted repeats play an important role in riboswitches, which are RNA regulatory elements that control the expression of genes that produce the mRNA, of which they are part.[10] A simplified example of the flavin mononucleotide (FMN) riboswitch is shown in the illustration. This riboswitch exists in the mRNA transcript and has several stem-loop structures upstream from the coding region. However, only the key stem-loops are shown in the illustration, which has been greatly simplified to help show the role of the inverted repeats. There are multiple inverted repeats in this riboswitch as indicated in green (yellow background) and blue (orange background).

In the absence of FMN, the Anti-termination structure is the preferred conformation for the mRNA transcript. It is created by base-pairing of the inverted repeat region circled in red. When FMN is present, it may bind to the loop and prevent formation of the Anti-termination structure. This allows two different sets of inverted repeats to base-pair and form the Termination structure.[20] The stem-loop on the 3' end is a transcriptional terminator because the sequence immediately following it is a string of uracils (U). If this stem-loop forms (due to the presence of FMN) as the growing RNA strand emerges from the RNA polymerase complex, it will create enough structural tension to cause the RNA strand to dissociate and thus terminate transcription. The dissociation occurs easily because the base-pairing between the U's in the RNA and the A's in the template strand are the weakest of all base-pairings.[10] Thus, at higher concentration levels, FMN down-regulates its own transcription by increasing the formation of the termination structure.
Mutations and disease
[edit]Inverted repeats are often described as "hotspots" of eukaryotic and prokaryotic genomic instability.[7] Long inverted repeats are deemed to greatly influence the stability of the genome of various organisms.[21] This is exemplified in E. coli, where genomic sequences with long inverted repeats are seldom replicated, but rather deleted with rapidity.[21] Again, the long inverted repeats observed in yeast greatly favor recombination within the same and adjacent chromosomes, resulting in an equally very high rate of deletion.[21] Finally, a very high rate of deletion and recombination were also observed in mammalian chromosomes regions with inverted repeats.[21] Reported differences in the stability of genomes of interrelated organisms are always an indication of a disparity in inverted repeats.[11] The instability results from the tendency of inverted repeats to fold into hairpin- or cruciform-like DNA structures. These special structures can hinder or confuse DNA replication and other genomic activities.[7] Thus, inverted repeats lead to special configurations in both RNA and DNA that can ultimately cause mutations and disease.[9]

The illustration shows an inverted repeat undergoing cruciform extrusion. DNA in the region of the inverted repeat unwinds and then recombines, forming a four-way junction with two stem-loop structures. The cruciform structure occurs because the inverted repeat sequences self-pair to each other on their own strand.[22]
Extruded cruciforms can lead to frameshift mutations when a DNA sequence has inverted repeats in the form of a palindrome combined with regions of direct repeats on either side. During transcription, slippage and partial dissociation of the polymerase from the template strand can lead to both deletion and insertion mutations.[9] Deletion occurs when a portion of the unwound template strand forms a stem-loop that gets "skipped" by the transcription machinery. Insertion occurs when a stem-loop forms in a dissociated portion of the nascent (newly synthesized) strand causing a portion of the template strand to be transcribed twice.[9]
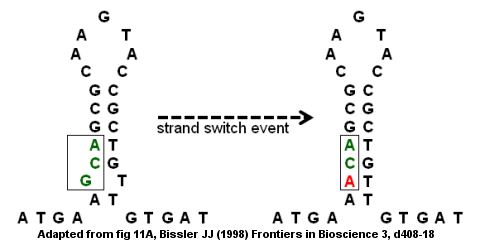
Antithrombin deficiency from a point mutation
[edit]Imperfect inverted repeats can lead to mutations through intrastrand and interstrand switching.[9] The antithrombin III gene's coding region is an example of an imperfect inverted repeat as shown in the figure on the right. The stem-loop structure forms with a bump at the bottom because the G and T do not pair up. A strand switch event could result in the G (in the bump) being replaced by an A which removes the "imperfection" in the inverted repeat and provides a stronger stem-loop structure. However, the replacement also creates a point mutation converting the GCA codon to ACA. If the strand switch event is followed by a second round of DNA replication, the mutation may become fixed in the genome and lead to disease. Specifically, the missense mutation would lead to a defective gene and a deficiency in antithrombin which could result in the development of venous thromboembolism (blood clots within a vein).[9]

Osteogenesis imperfecta from a frameshift mutation
[edit]Mutations in the collagen gene can lead to the disease Osteogenesis Imperfecta, which is characterized by brittle bones.[9] In the illustration, a stem-loop formed from an imperfect inverted repeat is mutated with a thymine (T) nucleotide insertion as a result of an inter- or intrastrand switch. The addition of the T creates a base-pairing "match up" with the adenine (A) that was previously a "bump" on the left side of the stem. While this addition makes the stem stronger and perfects the inverted repeat, it also creates a frameshift mutation in the nucleotide sequence which alters the reading frame and will result in an incorrect expression of the gene.[9]
Programs and databases
[edit]The following list provides information and external links to various programs and databases for inverted repeats:
- non-B DB A Database for Integrated Annotations and Analysis of non-B DNA Forming Motifs.[23] This database is provided by The Advanced Biomedical Computing Center (ABCC) at then Frederick National Laboratory for Cancer Research (FNLCR). It covers the A-DNA and Z-DNA conformations otherwise known as "non-B DNAs" because they are not the more common B-DNA form of a right-handed Watson-Crick double-helix. These "non-B DNAs" include left-handed Z-DNA, cruciform, triplex, tetraplex and hairpin structures.[23] Searches can be performed on a variety of "repeat types" (including inverted repeats) and on several species.
- Inverted Repeats Database Archived 2020-09-01 at the Wayback Machine Boston University. This database is a web application that allows query and analysis of repeats held in the PUBLIC DATABASE project. Scientists can also analyze their own sequences with the Inverted Repeats Finder algorithm.[24]
- P-MITE: a Plant MITE database — this database for Miniature Inverted-repeat Transposable Elements (MITEs) contains sequences from plant genomes. Sequences may be searched or downloaded from the database.[25]
- EMBOSS is the "European Molecular Biology Open Software Suite" which runs on UNIX and UNIX-like operating systems.[26] Documentation and program source files are available on the EMBOSS website. Applications specifically related to inverted repeats are listed below:
- EMBOSS einverted: Finds inverted repeats in nucleotide sequences. Threshold values can be set to limit the scope of the search.[26]
- EMBOSS palindrome: Finds palindromes such as stem loop regions in nucleotide sequences. The program will find sequences that include sections of mismatches and gaps that may correspond to bulges in a stem loop.[26]
References
[edit]- ^ a b c d Ussery, David W.; Wassenaar, Trudy; Borini, Stefano (2008-12-22). "Word Frequencies, Repeats, and Repeat-related Structures in Bacterial Genomes". Computing for Comparative Microbial Genomics: Bioinformatics for Microbiologists. Computational Biology. Vol. 8 (1 ed.). Springer. pp. 133–144. ISBN 978-1-84800-254-8.
- ^ Ye, Congting; Ji, Guoli; Liang, Chun (2014). "detectIR: A Novel Program for Detecting Perfect and Imperfect Inverted Repeats Using Complex Numbers and Vector Calculation". PLOS ONE. 9 (11) e113349. Bibcode:2014PLoSO...9k3349Y. doi:10.1371/journal.pone.0113349. PMC 4237412. PMID 25409465.
- ^ a b Richards, GR; Richards, RI (Apr 25, 1995). "Simple tandem DNA repeats and human genetic disease". Proceedings of the National Academy of Sciences of the United States of America. 92 (9): 3636–41. Bibcode:1995PNAS...92.3636S. doi:10.1073/pnas.92.9.3636. PMC 42017. PMID 7731957.
- ^ van Belkum, A; Scherer, S; van Alphen, L; Verbrugh, H (June 1998). "Short-sequence DNA repeats in prokaryotic genomes". Microbiology and Molecular Biology Reviews. 62 (2): 275–93. doi:10.1128/MMBR.62.2.275-293.1998. PMC 98915. PMID 9618442.
- ^ a b Ramel, C (June 1997). "Mini- and microsatellites". Environmental Health Perspectives. 105 (Suppl 4): 781–9. doi:10.2307/3433284. JSTOR 3433284. PMC 1470042. PMID 9255562.
- ^ a b Eichler, EE (August 1998). "Masquerading repeats: paralogous pitfalls of the human genome". Genome Research. 8 (8): 758–62. doi:10.1101/gr.8.8.758. PMID 9724321.
- ^ a b c Mirkin, I; Narayanan, V; Lobachev, KS; Mirkin, SM (Jul 22, 2008). "Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins". Proceedings of the National Academy of Sciences of the United States of America. 105 (29): 9936–41. Bibcode:2008PNAS..105.9936V. doi:10.1073/pnas.0804510105. PMC 2481305. PMID 18632578.
- ^ Lin, CT; Lin, WH; Lyu, YL; Whang-Peng, J (Sep 1, 2001). "Inverted repeats as genetic elements for promoting DNA inverted duplication: implications in gene amplification". Nucleic Acids Research. 29 (17): 3529–38. doi:10.1093/nar/29.17.3529. PMC 55881. PMID 11522822.
- ^ a b c d e f g h Bissler, JJ (Mar 27, 1998). "DNA inverted repeats and human disease" (PDF). Frontiers in Bioscience. 3 (4) A284: d408–18. doi:10.2741/a284. PMID 9516381. S2CID 12982. Archived from the original (PDF) on March 3, 2019.
- ^ a b c d e f g School, James D. Watson, Cold Spring Harbor Laboratory, Tania A. Baker, Massachusetts Institute of Technology, Stephen P. Bell, Massachusetts Institute of Technology, Alexander Gann, Cold Spring Harbor Laboratory, Michael Levine, University of California, Berkeley, Richard Losik, Harvard University; with Stephen C. Harrison, Harvard Medical (2014). Molecular biology of the gene (Seventh ed.). Boston: Benjamin-Cummings Publishing Company. ISBN 978-0-321-76243-6.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ a b Achaz, G; Coissac, E; Netter, P; Rocha, EP (August 2003). "Associations between inverted repeats and the structural evolution of bacterial genomes". Genetics. 164 (4): 1279–89. doi:10.1093/genetics/164.4.1279. PMC 1462642. PMID 12930739.
- ^ Zhang, HH; Xu, HE; Shen, YH; Han, MJ; Zhang, Z (January 2013). "The Origin and Evolution of Six Miniature Inverted-Repeat Transposable Elements in Bombyx mori and Rhodnius prolixus". Genome Biology and Evolution. 5 (11): 2020–31. doi:10.1093/gbe/evt153. PMC 3845634. PMID 24115603.
- ^ a b Pearson, CE; Zorbas, H; Price, GB; Zannis-Hadjopoulos, M (October 1996). "Inverted repeats, stem-loops, and cruciforms: significance for initiation of DNA replication". Journal of Cellular Biochemistry. 63 (1): 1–22. doi:10.1002/(SICI)1097-4644(199610)63:1<1::AID-JCB1>3.0.CO;2-3. PMID 8891900. S2CID 22204780.
- ^ a b c Heringa, J (June 1998). "Detection of internal repeats: how common are they?". Current Opinion in Structural Biology. 8 (3): 338–45. doi:10.1016/S0959-440X(98)80068-7. PMID 9666330.
- ^ Treangen, TJ; Abraham, AL; Touchon, M; Rocha, EP (May 2009). "Genesis, effects and fates of repeats in prokaryotic genomes" (PDF). FEMS Microbiology Reviews. 33 (3): 539–71. doi:10.1111/j.1574-6976.2009.00169.x. PMID 19396957.
- ^ Ladoukakis, ED; Eyre-Walker, A (September 2008). "The excess of small inverted repeats in prokaryotes" (PDF). Journal of Molecular Evolution. 67 (3): 291–300. Bibcode:2008JMolE..67..291L. CiteSeerX 10.1.1.578.7466. doi:10.1007/s00239-008-9151-z. PMID 18696026. S2CID 29953202.
- ^ Hosseini, M; Pratas, D; Pinho, AJ (2017). "On the Role of Inverted Repeats in DNA Sequence Similarity". 11th International Conference on Practical Applications of Computational Biology & Bioinformatics. Advances in Intelligent Systems and Computing. Vol. 616. Springer. pp. 228–236. doi:10.1007/978-3-319-60816-7_28. ISBN 978-3-319-60815-0.
- ^ a b Staple, DW; Butcher, SE (June 2005). "Pseudoknots: RNA structures with diverse functions". PLOS Biology. 3 (6) e213. doi:10.1371/journal.pbio.0030213. PMC 1149493. PMID 15941360.

- ^ a b Chen, JL; Greider, CW (Jun 7, 2005). "Functional analysis of the pseudoknot structure in human telomerase RNA". Proceedings of the National Academy of Sciences of the United States of America. 102 (23): 8080–5, discussion 8077–9. Bibcode:2005PNAS..102.8080C. doi:10.1073/pnas.0502259102. PMC 1149427. PMID 15849264.
- ^ Winkler, WC; Cohen-Chalamish, S; Breaker, RR (Dec 10, 2002). "An mRNA structure that controls gene expression by binding FMN". Proceedings of the National Academy of Sciences of the United States of America. 99 (25): 15908–13. Bibcode:2002PNAS...9915908W. doi:10.1073/pnas.212628899. PMC 138538. PMID 12456892.
- ^ a b c d Stormo, G; Chang, KY; Varley, K; Stormo, GD (Feb 28, 2007). Hall, Neil (ed.). "Evidence for active maintenance of inverted repeat structures identified by a comparative genomic approach". PLOS ONE. 2 (2) e262. Bibcode:2007PLoSO...2..262Z. doi:10.1371/journal.pone.0000262. PMC 1803023. PMID 17327921.

- ^ Ramreddy, T; Sachidanandam, R; Strick, TR (May 2011). "Real-time detection of cruciform extrusion by single-molecule DNA nanomanipulation". Nucleic Acids Research. 39 (10): 4275–83. doi:10.1093/nar/gkr008. PMC 3105387. PMID 21266478.
- ^ a b Cer, RZ; Donohue, DE; Mudunuri, US; Temiz, NA; Loss, MA; Starner, NJ; Halusa, GN; Volfovsky, N; Yi, M; Luke, BT; Bacolla, A; Collins, JR; Stephens, RM (January 2013). "Non-B DB v2.0: a database of predicted non-B DNA-forming motifs and its associated tools". Nucleic Acids Research. 41 (Database issue): D94 – D100. doi:10.1093/nar/gks955. PMC 3531222. PMID 23125372.
- ^ Gelfand, Y; Rodriguez, A; Benson, G (January 2007). "TRDB--the Tandem Repeats Database". Nucleic Acids Research. 35 (Database issue): D80–7. doi:10.1093/nar/gkl1013. PMC 1781109. PMID 17175540.
- ^ Chen, J; Hu, Q; Zhang, Y; Lu, C; Kuang, H (Oct 29, 2013). "P-MITE: a database for plant miniature inverted-repeat transposable elements". Nucleic Acids Research. 42 (1): D1176–81. doi:10.1093/nar/gkt1000. PMC 3964958. PMID 24174541.
- ^ a b c Rice, P; Longden, I; Bleasby, A (June 2000). "EMBOSS: the European Molecular Biology Open Software Suite". Trends in Genetics. 16 (6): 276–7. doi:10.1016/S0168-9525(00)02024-2. PMID 10827456.
External links
[edit]- Inverted+Repeat+Sequence at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
Inverted repeat
View on GrokipediaDefinition and Structure
Core Definition
An inverted repeat (IR) is a sequence motif in nucleic acids consisting of a segment of nucleotides followed downstream by its reverse complement, which may be adjacent or separated by a spacer sequence.[4] This arrangement allows the two halves to base-pair with each other, potentially forming stable secondary structures.[5] Inverted repeats occur in both DNA and RNA, where they contribute to various regulatory and structural roles in molecular biology.[6] Inverted repeats can be classified as perfect or imperfect based on the degree of sequence complementarity between the two arms. Perfect inverted repeats exhibit exact matching between the sequence and its reverse complement, enabling complete base pairing without mismatches.[7] Imperfect inverted repeats, in contrast, contain one or more mismatches, insertions, or deletions, resulting in partial complementarity.[7] A common notation for an inverted repeat is 5'-XYZ...Z'Y'X'-3', where the first half (XYZ...) is followed by the reverse complement (Z'Y'X'...) of that sequence, with primes denoting the complemented bases.[4] In DNA, inverted repeats can extrude into hairpin or cruciform structures, particularly under conditions of negative supercoiling, where the two arms pair to form a stem with a loop or junction.[8] In RNA, these motifs typically fold into stem-loop structures, with the paired arms creating a double-stranded stem and the spacer or mismatched region forming an unpaired loop.[9]Structural Characteristics
An inverted repeat consists of two arms composed of repeated nucleotide sequences that are reverse complements of each other, separated by an intervening spacer region of non-repeated sequence.[10] The arms enable intrastrand base pairing, while the spacer typically remains unpaired and can form a loop in the folded structure.[11] Inverted repeats exhibit significant length variations, with short forms ranging from 2-6 base pairs (often termed micro-inverted repeats) to longer ones exceeding 30 base pairs, and extended versions reaching tens of kilobases, as seen in the large inverted repeats of many chloroplast genomes.[12][13] Imperfect inverted repeats incorporate mismatches, insertions, or bulges within the arms, reducing complementarity but still allowing partial base pairing.[10] These motifs possess the potential to fold into secondary structures through Watson-Crick base pairing between the arms, forming cruciforms in double-stranded DNA or hairpins in single-stranded RNA.[14][15] The thermodynamic stability of such structures is determined by the Gibbs free energy change (ΔG), primarily through nearest-neighbor interactions and loop contributions, with more negative ΔG indicating greater stability.[16] Detection of inverted repeats generally requires a minimum arm length of 5-10 nucleotides to ensure stable pairing, with maximum spacer lengths limited to under 50 nucleotides for efficient folding and hybridization.[17][11]Illustrative Examples
A simple example of an inverted repeat in DNA is the sequence 5'-GAATTC...CTTAAG-3', where "GAATTC" is followed by its reverse complement "CTTAAG" after a spacer region; this illustrates the core principle of an inverted repeat as one segment being the reverse complement of the other.[1] In RNA, an inverted repeat can form a stem-loop structure, such as 5'-GCAAGC...GCUUCG-3', where "GCAAGC" pairs with its reverse complement "GCUUCG" via base pairing in the folded form, demonstrating how such sequences enable intramolecular hybridization.[18] A longer example occurs in the inverted terminal repeats (ITRs) of adenovirus genomes, which consist of approximately 100-140 base pairs at both ends of the linear DNA molecule, with each end being the reverse complement of the other to facilitate circularization or replication initiation.[19] To visualize an inverted repeat, consider its linear form as a single-stranded sequence like 5'-ABC...CBA-3', where "CBA" is the reverse complement of "ABC"; in the folded form, it adopts a double-stranded stem with a loop in the spacer, represented schematically as:5'- A B C -3'
| | |
3'- T S G A B C -5' (loop omitted for simplicity)
5'- A B C -3'
| | |
3'- T S G A B C -5' (loop omitted for simplicity)
