Recent from talks
All channels
Be the first to start a discussion here.
Be the first to start a discussion here.
Be the first to start a discussion here.
Be the first to start a discussion here.
Welcome to the community hub built to collect knowledge and have discussions related to RNA world.
Nothing was collected or created yet.
RNA world
View on Wikipediafrom Wikipedia
Not found
RNA world
View on Grokipediafrom Grokipedia
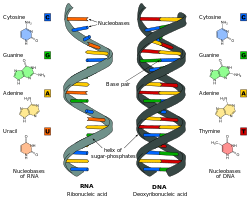
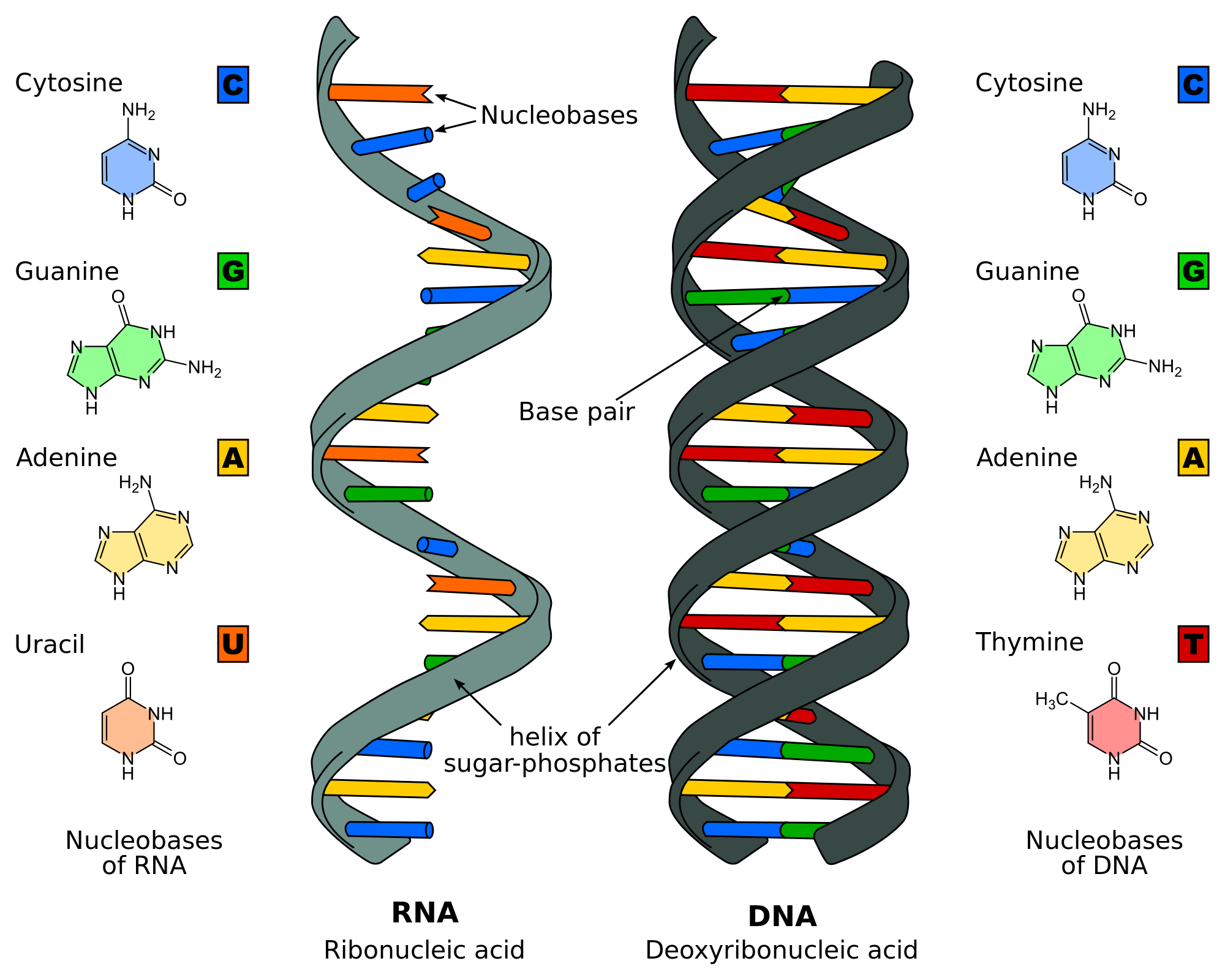
The RNA world hypothesis posits that early life on Earth evolved from a stage where RNA molecules served dual roles as both genetic material, storing and transmitting information via self-replication, and as catalysts, performing biochemical reactions akin to enzymes, prior to the emergence of DNA and proteins.[1] This model suggests that RNA's versatility allowed it to bootstrap the development of more complex cellular machinery, with genetic continuity maintained through RNA replication driven by Watson-Crick base-pairing.[1]
The concept traces its roots to the 1960s, when researchers including Francis Crick, Carl Woese, and Leslie Orgel independently proposed that RNA or a similar nucleic acid could have predated proteins in primordial biochemistry.[1] The term "RNA world" was coined by Walter Gilbert in 1986, building on these ideas to describe a prebiotic era dominated by RNA.[2] A pivotal advancement came in 1982–1983 with the discovery of ribozymes—RNA molecules with catalytic activity—by Thomas Cech, who identified self-splicing introns in Tetrahymena, and Sidney Altman, who characterized the RNA component of RNase P as an enzyme.[3] This breakthrough, for which Cech and Altman shared the 1989 Nobel Prize in Chemistry, provided direct evidence that RNA could function catalytically, challenging the protein-centric view of enzymology.
Supporting evidence includes the central role of RNA in modern biology, such as the ribosome's peptidyl transferase center, which is a ribozyme responsible for protein synthesis.[1] Laboratory experiments have demonstrated RNA polymerase ribozymes capable of replicating other RNA strands, as shown by Bartel and Szostak in 1993, and continuous in vitro evolution of self-replicating RNA systems by Lincoln and Joyce in 2009; more recently, in 2024, researchers at the Salk Institute developed high-fidelity RNA polymerase ribozymes enabling molecular-scale Darwinian evolution.[1][4] Prebiotic chemistry studies further bolster the hypothesis, with demonstrations of nucleotide synthesis and non-enzymatic RNA polymerization under simulated early Earth conditions.[5]
Despite its prominence, the RNA world faces challenges, including the instability of RNA in prebiotic environments and the difficulty of achieving robust, error-free replication without protein assistance.[5] Recent models propose hybrid scenarios, such as an RNA-DNA world transition involving reverse transcriptase-like ribozymes to incorporate DNA for greater stability.[6] Ongoing research continues to refine the hypothesis through advances in synthetic biology and astrobiology, aiming to reconstruct plausible pathways for life's origins.[5]