Recent from talks
Nothing was collected or created yet.
Motor protein
View on Wikipedia
Motor proteins are a class of molecular motors that can move along the cytoskeleton of cells. They do this by converting chemical energy into mechanical work by the hydrolysis of ATP.[1]
Cellular functions
[edit]
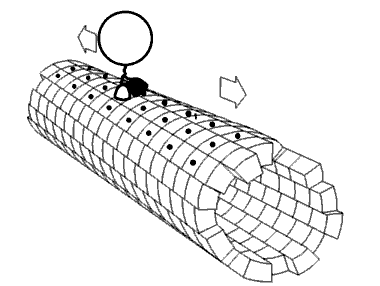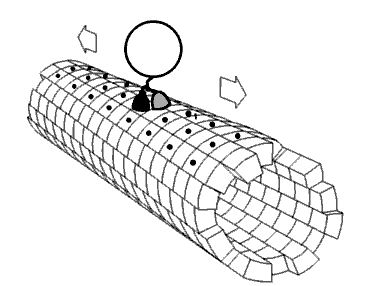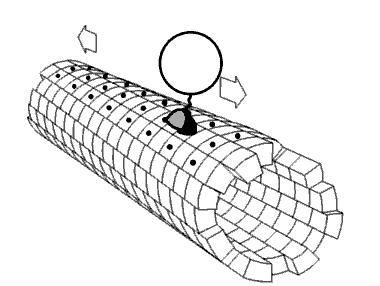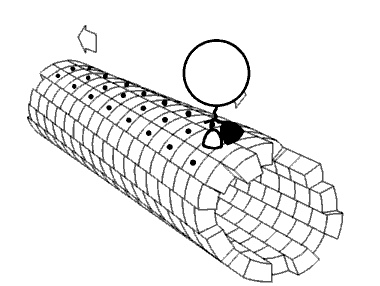
Motor proteins are the driving force behind most active transport of proteins and vesicles in the cytoplasm. Kinesins and cytoplasmic dyneins play essential roles in intracellular transport such as axonal transport and in the formation of the spindle apparatus and the separation of the chromosomes during mitosis and meiosis. Axonemal dynein, found in cilia and flagella, is crucial to cell motility in spermatozoa, and fluid transport in trachea.[citation needed] The muscle protein myosin "motors" the contraction of muscle fibers in animals.
Diseases associated with motor protein defects
[edit]The importance of motor proteins in cells becomes evident when they fail to fulfill their function. For example, kinesin deficiencies have been identified as the cause for Charcot-Marie-Tooth disease and some kidney diseases. Dynein deficiencies can lead to chronic infections of the respiratory tract as cilia fail to function without dynein. Numerous myosin deficiencies are related to disease states and genetic syndromes. Because myosin II is essential for muscle contraction, defects in muscular myosin predictably cause myopathies. Myosin is necessary in the process of hearing because of its role in the growth of stereocilia so defects in myosin protein structure can lead to Usher syndrome and non-syndromic deafness.[2]
Cytoskeletal motor proteins
[edit]Motor proteins utilizing the cytoskeleton for movement fall into two categories based on their substrate: microfilaments or microtubules. Actin-based motor proteins (myosin) move along microfilaments through interaction with actin, and microtubule motors (dynein and kinesin) move along microtubules through interaction with tubulin.[citation needed]
There are two basic types of microtubule motors: plus-end motors and minus-end motors, depending on the direction in which they "walk" along the microtubule cables within the cell.
Actin motors
[edit]Myosin
[edit]Myosins are a superfamily of actin motor proteins that convert chemical energy in the form of ATP to mechanical energy, thus generating force and movement. The first identified myosin, myosin II, is responsible for generating muscle contraction. Myosin II is an elongated protein that is formed from two heavy chains with motor heads and two light chains. Each myosin head contains actin and ATP binding site. The myosin heads bind and hydrolyze ATP, which provides the energy to walk toward the plus end of an actin filament. Myosin II are also vital in the process of cell division. For example, non-muscle myosin II bipolar thick filaments provide the force of contraction needed to divide the cell into two daughter cells during cytokinesis. In addition to myosin II, many other myosin types are responsible for variety of movement of non-muscle cells. For example, myosin is involved in intracellular organization and the protrusion of actin-rich structures at the cell surface. Myosin V is involved in vesicle and organelle transport.[3][4] Myosin XI is involved in cytoplasmic streaming, wherein movement along microfilament networks in the cell allows organelles and cytoplasm to stream in a particular direction.[5] Eighteen different classes of myosins are known.[6]
Genomic representation of myosin motors:[7]
- Fungi (yeast): 5
- Plants (Arabidopsis): 17
- Insects (Drosophila): 13
- Mammals (human): 40
- Chromadorea ( nematode C. elegans): 15
Microtubule motors
[edit]Kinesin
[edit]Kinesins are a superfamily of related motor proteins that use a microtubule track in anterograde movement. They are vital to spindle formation in mitotic and meiotic chromosome separation during cell division and are also responsible for shuttling mitochondria, Golgi bodies, and vesicles within eukaryotic cells. Kinesins have two heavy chains and two light chains per active motor. The two globular head motor domains in heavy chains can convert the chemical energy of ATP hydrolysis into mechanical work to move along microtubules.[8] The direction in which cargo is transported can be towards the plus-end or the minus-end, depending on the type of kinesin. In general, kinesins with N-terminal motor domains move their cargo towards the plus ends of microtubules located at the cell periphery, while kinesins with C-terminal motor domains move cargo towards the minus ends of microtubules located at the nucleus. Fourteen distinct kinesin families are known, with some additional kinesin-like proteins that cannot be classified into these families.[9]
Genomic representation of kinesin motors:[7]
- Fungi (yeast): 6
- Plants (Arabidopsis thaliana): 61
- Insects (Drosophila melanogaster): 25
- Mammals (human): 45
Dynein
[edit]Dyneins are microtubule motors capable of a retrograde sliding movement. Dynein complexes are much larger and more complex than kinesin and myosin motors. Dyneins are composed of two or three heavy chains and a large and variable number of associated light chains. Dyneins drive intracellular transport toward the minus end of microtubules which lies in the microtubule organizing center near the nucleus.[10] The dynein family has two major branches. Axonemal dyneins facilitate the beating of cilia and flagella by rapid and efficient sliding movements of microtubules. Another branch is cytoplasmic dyneins which facilitate the transport of intracellular cargos. Compared to 15 types of axonemal dynein, only two cytoplasmic forms are known.[11]
Genomic representation of dynein motors:[7]
- Fungi (yeast): 1
- Plants (Arabidopsis thaliana): 0
- Insects (Drosophila melanogaster): 13
- Mammals (human): 14-15
Plant-specific motors
[edit]In contrast to animals, fungi and non-vascular plants, the cells of flowering plants lack dynein motors. However, they contain a larger number of different kinesins. Many of these plant-specific kinesin groups are specialized for functions during plant cell mitosis.[12] Plant cells differ from animal cells in that they have a cell wall. During mitosis, the new cell wall is built by the formation of a cell plate starting in the center of the cell. This process is facilitated by a phragmoplast, a microtubule array unique to plant cell mitosis. The building of cell plate and ultimately the new cell wall requires kinesin-like motor proteins.[13]
Another motor protein essential for plant cell division is kinesin-like calmodulin-binding protein (KCBP), which is unique to plants and part kinesin and part myosin.[14]
Other molecular motors
[edit]Besides the motor proteins above, there are many more types of proteins capable of generating forces and torque in the cell. Many of these molecular motors are ubiquitous in both prokaryotic and eukaryotic cells, although some, such as those involved with cytoskeletal elements or chromatin, are unique to eukaryotes. The motor protein prestin,[15] expressed in mammalian cochlear outer hair cells, produces mechanical amplification in the cochlea. It is a direct voltage-to-force converter, which operates at the microsecond rate and possesses piezoelectric properties.
See also
[edit]References
[edit]- ^ Alberts, Bruce; Johnson, Alexander; Lewis, Julian; Raff, Martin; Roberts, Keith; Walter, Peter (2002), "Molecular Motors", Molecular Biology of the Cell. 4th edition, Garland Science, retrieved 2025-04-03
- ^ Hirokawa N, Takemura R (October 2003). "Biochemical and molecular characterization of diseases linked to motor proteins". Trends in Biochemical Sciences. 28 (10): 558–65. doi:10.1016/j.tibs.2003.08.006. PMID 14559185.
- ^ Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002-01-01). "Molecular Motors". NCBI - National Institutes of Health.
- ^ Warshaw, DM (February 2012). "Tilting and twirling as myosin V steps along actin filaments as detected by fluorescence polarization". The Journal of General Physiology. 139 (2): 97–100. doi:10.1085/jgp.201210769. PMC 3269787. PMID 22291143.
- ^ Hartman MA, Spudich JA (April 2012). "The myosin superfamily at a glance". Journal of Cell Science. 125 (Pt 7): 1627–32. doi:10.1242/jcs.094300. PMC 3346823. PMID 22566666.
- ^ Thompson RF, Langford GM (November 2002). "Myosin superfamily evolutionary history". The Anatomical Record. 268 (3): 276–89. doi:10.1002/ar.10160. PMID 12382324. S2CID 635349.
- ^ a b c Vale RD (February 2003). "The molecular motor toolbox for intracellular transport". Cell. 112 (4): 467–80. doi:10.1016/S0092-8674(03)00111-9. PMID 12600311.
- ^ Verhey KJ, Kaul N, Soppina V (2011-01-01). "Kinesin assembly and movement in cells". Annual Review of Biophysics. 40: 267–88. doi:10.1146/annurev-biophys-042910-155310. PMID 21332353.
- ^ Miki H, Okada Y, Hirokawa N (September 2005). "Analysis of the kinesin superfamily: insights into structure and function". Trends in Cell Biology. 15 (9): 467–76. doi:10.1016/j.tcb.2005.07.006. PMID 16084724.
- ^ Roberts AJ, Kon T, Knight PJ, Sutoh K, Burgess SA (November 2013). "Functions and mechanics of dynein motor proteins". Nature Reviews. Molecular Cell Biology. 14 (11): 713–26. doi:10.1038/nrm3667. PMC 3972880. PMID 24064538.
- ^ Mallik R, Gross SP (November 2004). "Molecular motors: strategies to get along". Current Biology. 14 (22): R971-82. Bibcode:2004CBio...14.R971M. doi:10.1016/j.cub.2004.10.046. PMID 15556858. S2CID 14240073.
- ^ Vanstraelen M, Inzé D, Geelen D (April 2006). "Mitosis-specific kinesins in Arabidopsis". Trends in Plant Science. 11 (4): 167–75. Bibcode:2006TPS....11..167V. doi:10.1016/j.tplants.2006.02.004. hdl:1854/LU-364298. PMID 16530461.
- ^ Smith LG (March 2002). "Plant cytokinesis: motoring to the finish". Current Biology. 12 (6): R206-8. Bibcode:2002CBio...12.R206S. doi:10.1016/S0960-9822(02)00751-0. PMID 11909547.
- ^ Abdel-Ghany SE, Day IS, Simmons MP, Kugrens P, Reddy AS (July 2005). "Origin and evolution of Kinesin-like calmodulin-binding protein". Plant Physiology. 138 (3): 1711–22. doi:10.1104/pp.105.060913. PMC 1176440. PMID 15951483.
- ^ Dallos P, Fakler B (February 2002). "Prestin, a new type of motor protein". Nature Reviews. Molecular Cell Biology. 3 (2): 104–11. doi:10.1038/nrm730. PMID 11836512. S2CID 7333228.
External links
[edit]- MBInfo - What are Motor Proteins?
- Ron Vale's Seminar: "Molecular Motor Proteins"
- Biology of Motor Proteins Institute for Biophysical Chemistry, Göttingen
- Jonathan Howard (2001), Mechanics of motor proteins and the cytoskeleton. ISBN 9780878933334