Recent from talks
Nothing was collected or created yet.
Spliceosome
View on WikipediaA spliceosome is a large ribonucleoprotein (RNP) complex found primarily within the nucleus of eukaryotic cells. The spliceosome is assembled from small nuclear RNAs (snRNA) and numerous proteins. Small nuclear RNA (snRNA) molecules bind to specific proteins to form a small nuclear ribonucleoprotein complex (snRNP, pronounced "snurps"), which in turn combines with other snRNPs to form a large ribonucleoprotein complex called a spliceosome. The spliceosome removes introns from a transcribed pre-mRNA, a type of primary transcript. This process is generally referred to as splicing.[1] An analogy is a film editor, who selectively cuts out irrelevant or incorrect material (equivalent to the introns) from the initial film and sends the cleaned-up version to the director for the final cut.[citation needed]
However, sometimes the RNA within the intron acts as a ribozyme, splicing itself without the use of a spliceosome or protein enzymes.[citation needed]
History
[edit]In 1977, work by the Sharp and Roberts labs revealed that genes of higher organisms are "split" or present in several distinct segments along the DNA molecule.[2][3] The coding regions of the gene are separated by non-coding DNA that is not involved in protein expression. The split gene structure was found when adenoviral mRNAs were hybridized to endonuclease cleavage fragments of single stranded viral DNA.[2] It was observed that the mRNAs of the mRNA-DNA hybrids contained 5' and 3' tails of non-hydrogen bonded regions. When larger fragments of viral DNAs were used, forked structures of looped out DNA were observed when hybridized to the viral mRNAs. It was realised that the looped out regions, the introns, are excised from the precursor mRNAs in a process Sharp named "splicing". The split gene structure was subsequently found to be common to most eukaryotic genes. Phillip Sharp and Richard J. Roberts were awarded the Nobel Prize in Medicine 1993 for the discovery of introns and the splicing process.
Composition
[edit]Each spliceosome is composed of five small nuclear RNAs (snRNA) and a range of associated protein factors. When these small RNAs are combined with the protein factors, they make RNA-protein complexes called snRNPs (small nuclear ribonucleoproteins, pronounced "snurps"). The snRNAs that make up the major spliceosome are named U1, U2, U4, U5, and U6, so-called because they are rich in uridine, and participate in several RNA-RNA and RNA-protein interactions.[1]
The assembly of the spliceosome occurs on each pre-mRNA (also known as heterogeneous nuclear RNA, hn-RNA) at each exon:intron junction. The pre-mRNA introns contains specific sequence elements that are recognized and utilized during spliceosome assembly. These include the 5' end splice site, the branch point sequence, the polypyrimidine tract, and the 3' end splice site. The spliceosome catalyzes the removal of introns, and the ligation of the flanking exons.[citation needed]
Introns typically have a GU nucleotide sequence at the 5' end splice site, and an AG at the 3' end splice site. The 3' splice site can be further defined by a variable length of polypyrimidines, called the polypyrimidine tract (PPT), which serves the dual function of recruiting factors to the 3' splice site and possibly recruiting factors to the branch point sequence (BPS). The BPS contains the conserved adenosine required for the first step of splicing.[citation needed]
Many proteins exhibit a zinc-binding motif, which underscores the importance of zinc in the splicing mechanism.[4][5][6] The first molecular-resolution reconstruction of U4/U6.U5 triple small nuclear ribonucleoprotein (tri-snRNP) complex was reported in 2016.[7]

Cryo-EM has been applied extensively by Shi et al. to elucidate the near-/atomic structure of spliceosome in both yeast[9] and humans.[10] The molecular framework of spliceosome at near-atomic-resolution demonstrates Spp42 component of U5 snRNP forms a central scaffold and anchors the catalytic center in yeast. The atomic structure of the human spliceosome illustrates the step II component Slu7 adopts an extended structure, poised for selection of the 3'-splice site. All five metals (assigned as Mg2+) in the yeast complex are preserved in the human complex.[citation needed]
Alternative splicing
[edit]Alternative splicing (the re-combination of different exons) is a major source of genetic diversity in eukaryotes. Splice variants have been used to account for the relatively small number of protein coding genes in the human genome, currently estimated at around 20,000. One particular Drosophila gene, Dscam, has been speculated to be alternatively spliced into 38,000 different mRNAs, assuming all of its exons can splice independently of each other.[11]
Location of splicing
[edit]Pre-mRNA splicing factors were originally found to be concentrated in nuclear bodies known as nuclear speckles.[12] It was originally postulated that nuclear speckles are either sites of mRNA splicing or storage sites of mRNA splicing factors. It is now understood that nuclear speckles help concentrate splicing factors near genes that are physically located close to them. Genes located farther from speckles can still be transcribed and spliced, but their splicing is less efficient compared to those closer to speckles.[13] RNA splicing is a biochemical reaction, and like all biochemical reactions, its rate depends on the concentration of enzymes and substrates. In this case, the enzymes are the spliceosomes, and the substrates are the pre-mRNAs. By varying the concentration of spliceosomes and pre-mRNAs based on their proximity to nuclear speckles, cells could potentially regulate the efficiency of splicing.[13]
Assembly
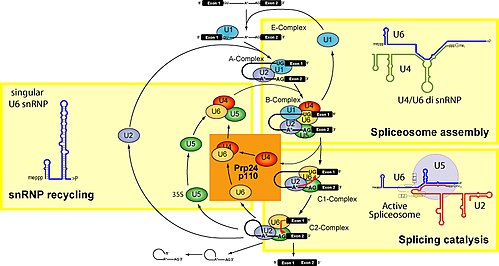
[edit]The model for formation of the spliceosome active site involves an ordered, stepwise assembly of discrete snRNP particles on the pre-mRNA substrate. The first recognition of pre-mRNAs involves U1 snRNP binding to the 5' end splice site of the pre-mRNA and other non-snRNP associated factors to form the commitment complex, or early (E) complex in mammals.[14][15] The commitment complex is an ATP-independent complex that commits the pre-mRNA to the splicing pathway.[16] U2 snRNP is recruited to the branch region through interactions with the E complex component U2AF (U2 snRNP auxiliary factor) and possibly U1 snRNP. In an ATP-dependent reaction, U2 snRNP becomes tightly associated with the branch point sequence (BPS) to form complex A. A duplex formed between U2 snRNP and the pre-mRNA branch region bulges out the branch adenosine specifying it as the nucleophile for the first transesterification.[17]
The presence of a pseudouridine residue in U2 snRNA, nearly opposite of the branch site, results in an altered conformation of the RNA-RNA duplex upon the U2 snRNP binding. Specifically, the altered structure of the duplex induced by the pseudouridine places the 2' OH of the bulged adenosine in a favorable position for the first step of splicing.[18] The U4/U5/U6 tri-snRNP (see Figure 1) is recruited to the assembling spliceosome to form complex B, and following several rearrangements, complex C is activated for catalysis.[19][20] It is unclear how the tri-snRNP is recruited to complex A, but this process may be mediated through protein-protein interactions and/or base pairing interactions between U2 snRNA and U6 snRNA.[citation needed]
The U5 snRNP interacts with sequences at the 5' and 3' splice sites via the invariant loop of U5 snRNA[21] and U5 protein components interact with the 3' splice site region.[22]
Upon recruitment of the tri-snRNP, several RNA-RNA rearrangements precede the first catalytic step and further rearrangements occur in the catalytically active spliceosome. Several of the RNA-RNA interactions are mutually exclusive; however, it is not known what triggers these interactions, nor the order of these rearrangements. The first rearrangement is probably the displacement of U1 snRNP from the 5' splice site and formation of a U6 snRNA interaction. It is known that U1 snRNP is only weakly associated with fully formed spliceosomes,[23] and U1 snRNP is inhibitory to the formation of a U6-5' splice site interaction on a model of substrate oligonucleotide containing a short 5' exon and 5' splice site.[24] Binding of U2 snRNP to the branch point sequence (BPS) is one example of an RNA-RNA interaction displacing a protein-RNA interaction. Upon recruitment of U2 snRNP, the branch binding protein SF1 in the commitment complex is displaced since the binding site of U2 snRNA and SF1 are mutually exclusive events.[citation needed]
Within the U2 snRNA, there are other mutually exclusive rearrangements that occur between competing conformations. For example, in the active form, stem loop IIa is favored; in the inactive form a mutually exclusive interaction between the loop and a downstream sequence predominates.[20] It is unclear how U4 is displaced from U6 snRNA, although RNA has been implicated in spliceosome assembly, and may function to unwind U4/U6 and promote the formation of a U2/U6 snRNA interaction. The interactions of U4/U6 stem loops I and II dissociate and the freed stem loop II region of U6 folds on itself to form an intramolecular stem loop and U4 is no longer required in further spliceosome assembly. The freed stem loop I region of U6 base pairs with U2 snRNA forming the U2/U6 helix I. However, the helix I structure is mutually exclusive with the 3' half of an internal 5' stem loop region of U2 snRNA.[citation needed]
Minor spliceosome
[edit]Some eukaryotes have a second spliceosome, the so-called minor spliceosome.[25] A group of less abundant snRNAs, U11, U12, U4atac, and U6atac, together with U5, are subunits of the minor spliceosome that splices a rare class of pre-mRNA introns, denoted U12-type. The minor spliceosome is located in the nucleus like its major counterpart,[26] though there are exceptions in some specialised cells including anucleate platelets[27] and the dendroplasm (dendrite cytoplasm) of neuronal cells.[28]
References
[edit]- ^ a b Will CL, Lührmann R (July 2011). "Spliceosome structure and function". Cold Spring Harbor Perspectives in Biology. 3 (7) a003707. doi:10.1101/cshperspect.a003707. PMC 3119917. PMID 21441581.
- ^ a b Berget SM, Moore C, Sharp PA (August 1977). "Spliced segments at the 5' terminus of adenovirus 2 late mRNA". Proceedings of the National Academy of Sciences of the United States of America. 74 (8): 3171–5. Bibcode:1977PNAS...74.3171B. doi:10.1073/pnas.74.8.3171. PMC 431482. PMID 269380.
- ^ Chow LT, Roberts JM, Lewis JB, Broker TR (August 1977). "A map of cytoplasmic RNA transcripts from lytic adenovirus type 2, determined by electron microscopy of RNA:DNA hybrids". Cell. 11 (4): 819–36. doi:10.1016/0092-8674(77)90294-X. PMID 890740. S2CID 37967144.
- ^ Agafonov DE, Deckert J, Wolf E, Odenwälder P, Bessonov S, Will CL, et al. (July 2011). "Semiquantitative proteomic analysis of the human spliceosome via a novel two-dimensional gel electrophoresis method". Molecular and Cellular Biology. 31 (13): 2667–82. doi:10.1128/mcb.05266-11. PMC 3133382. PMID 21536652.
- ^ Kuhn AN, van Santen MA, Schwienhorst A, Urlaub H, Lührmann R (January 2009). "Stalling of spliceosome assembly at distinct stages by small-molecule inhibitors of protein acetylation and deacetylation". RNA. 15 (1): 153–75. doi:10.1261/rna.1332609. PMC 2612777. PMID 19029308.
- ^ Patil V, Canzoneri JC, Samatov TR, Lührmann R, Oyelere AK (September 2012). "Molecular architecture of zinc chelating small molecules that inhibit spliceosome assembly at an early stage". RNA. 18 (9): 1605–11. doi:10.1261/rna.034819.112. PMC 3425776. PMID 22832025.
- ^ Cate JH (March 2016). "STRUCTURE. A Big Bang in spliceosome structural biology". Science. 351 (6280): 1390–2. doi:10.1126/science.aaf4465. PMID 27013712. S2CID 206648185.
- ^ Häcker I, Sander B, Golas MM, Wolf E, Karagöz E, Kastner B, et al. (November 2008). "Localization of Prp8, Brr2, Snu114 and U4/U6 proteins in the yeast tri-snRNP by electron microscopy". Nature Structural & Molecular Biology. 15 (11): 1206–12. doi:10.1038/nsmb.1506. PMID 18953335. S2CID 22982227.
- ^ Yan C, Hang J, Wan R, Huang M, Wong CC, Shi Y (September 2015). "Structure of a yeast spliceosome at 3.6-angstrom resolution". Science. 349 (6253): 1182–91. Bibcode:2015Sci...349.1182Y. doi:10.1126/science.aac7629. PMID 26292707. S2CID 22194712.
- ^ Zhang X, Yan C, Hang J, Finci LI, Lei J, Shi Y (May 2017). "An Atomic Structure of the Human Spliceosome". Cell. 169 (5): 918–929.e14. doi:10.1016/j.cell.2017.04.033. PMID 28502770.
- ^ Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, et al. (June 2000). "Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity". Cell. 101 (6): 671–84. doi:10.1016/S0092-8674(00)80878-8. PMID 10892653.
- ^ Spector DL, Lamond AI (Feb 2011). "Nuclear speckles". Review. Cold Spring Harbor Perspectives in Biology. 3 (2) a000646. doi:10.1101/cshperspect.a000646. PMC 3039535. PMID 20926517.
- ^ a b Bhat P, Chow A, Emert B, et al. (May 2024). "Genome organization around nuclear speckles drives mRNA splicing efficiency". Nature. 629 (5): 1165–1173. Bibcode:2024Natur.629.1165B. doi:10.1038/s41586-024-07429-6. PMC 11164319. PMID 38720076.
- ^ Jamison SF, Crow A, Garcia-Blanco MA (October 1992). "The spliceosome assembly pathway in mammalian extracts". Molecular and Cellular Biology. 12 (10): 4279–87. doi:10.1128/MCB.12.10.4279. PMC 360351. PMID 1383687.
- ^ Seraphin B, Rosbash M (October 1989). "Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing". Cell. 59 (2): 349–58. doi:10.1016/0092-8674(89)90296-1. PMID 2529976. S2CID 18553973.
- ^ Legrain P, Seraphin B, Rosbash M (September 1988). "Early commitment of yeast pre-mRNA to the spliceosome pathway". Molecular and Cellular Biology. 8 (9): 3755–60. doi:10.1128/MCB.8.9.3755. PMC 365433. PMID 3065622.
- ^ Query CC, Moore MJ, Sharp PA (March 1994). "Branch nucleophile selection in pre-mRNA splicing: evidence for the bulged duplex model". Genes & Development. 8 (5): 587–97. doi:10.1101/gad.8.5.587. PMID 7926752.
- ^ Newby MI, Greenbaum NL (December 2002). "Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine". Nature Structural Biology. 9 (12): 958–65. doi:10.1038/nsb873. PMID 12426583. S2CID 39628664.
- ^ Burge CB, Tuschl T, Sharp PA (1999). "Splicing precursors to mRNAs by the spliceosomes". In Gesteland RF, Cech TR, Atkins JF (eds.). The RNA World. Cold Spring Harbor Lab. Press. pp. 525–60. ISBN 978-0-87969-380-0.
- ^ a b Staley JP, Guthrie C (February 1998). "Mechanical devices of the spliceosome: motors, clocks, springs, and things". Cell. 92 (3): 315–26. doi:10.1016/S0092-8674(00)80925-3. PMID 9476892.
- ^ Newman AJ, Teigelkamp S, Beggs JD (November 1995). "snRNA interactions at 5' and 3' splice sites monitored by photoactivated crosslinking in yeast spliceosomes". RNA. 1 (9): 968–80. PMC 1369345. PMID 8548661. Archived from the original on 2005-02-23. Retrieved 2008-03-07.
- ^ Chiara MD, Palandjian L, Feld Kramer R, Reed R (August 1997). "Evidence that U5 snRNP recognizes the 3' splice site for catalytic step II in mammals". The EMBO Journal. 16 (15): 4746–59. doi:10.1093/emboj/16.15.4746. PMC 1170101. PMID 9303319.
- ^ Moore MJ, Sharp PA (September 1993). "Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing". Nature. 365 (6444): 364–8. Bibcode:1993Natur.365..364M. doi:10.1038/365364a0. PMID 8397340. S2CID 4361512.
- ^ Konforti BB, Koziolkiewicz MJ, Konarska MM (December 1993). "Disruption of base pairing between the 5' splice site and the 5' end of U1 snRNA is required for spliceosome assembly". Cell. 75 (5): 863–73. doi:10.1016/0092-8674(93)90531-T. PMID 8252623.
- ^ Patel AA, Steitz JA (December 2003). "Splicing double: insights from the second spliceosome". Nature Reviews. Molecular Cell Biology. 4 (12): 960–70. doi:10.1038/nrm1259. PMID 14685174. S2CID 21816910.
- ^ Pessa HK, Will CL, Meng X, Schneider C, Watkins NJ, Perälä N, et al. (June 2008). "Minor spliceosome components are predominantly localized in the nucleus". Proceedings of the National Academy of Sciences of the United States of America. 105 (25): 8655–60. doi:10.1073/pnas.0803646105. PMC 2438382. PMID 18559850.
- ^ Denis MM, Tolley ND, Bunting M, Schwertz H, Jiang H, Lindemann S, et al. (August 2005). "Escaping the nuclear confines: signal-dependent pre-mRNA splicing in anucleate platelets". Cell. 122 (3): 379–91. doi:10.1016/j.cell.2005.06.015. PMC 4401993. PMID 16096058.
- ^ Glanzer J, Miyashiro KY, Sul JY, Barrett L, Belt B, Haydon P, et al. (November 2005). "RNA splicing capability of live neuronal dendrites". Proceedings of the National Academy of Sciences of the United States of America. 102 (46): 16859–64. Bibcode:2005PNAS..10216859G. doi:10.1073/pnas.0503783102. PMC 1277967. PMID 16275927.
Further reading
[edit]- Butcher SE (2011). "Chapter 8. The Spliceosome and Its Metal Ions". In Sigel A, Sigel H, Sigel RK (eds.). Structural and catalytic roles of metal ions in RNA. Metal Ions in Life Sciences. Vol. 9. RSC Publishing. pp. 235–51. doi:10.1039/9781849732512-00235. ISBN 978-1-84973-094-5. PMID 22010274.
- Nilsen TW (December 2003). "The spliceosome: the most complex macromolecular machine in the cell?". BioEssays. 25 (12): 1147–9. doi:10.1002/bies.10394. PMID 14635248.
External links
[edit]- 3D macromolecular structures of Spliceosomes from the EM Data Bank(EMDB)
- Spliceosomes at the U.S. National Library of Medicine Medical Subject Headings (MeSH)

