Recent from talks
Nothing was collected or created yet.
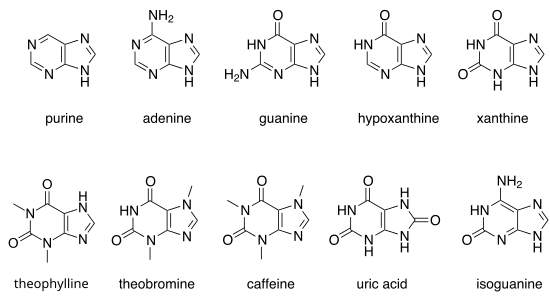
Purine
View on Wikipedia
 | |
 | |
 | |
| Names | |
|---|---|
| IUPAC name
9H-purine
| |
| Identifiers | |
3D model (JSmol)
|
|
| ChEBI | |
| ChEMBL | |
| ChemSpider | |
| ECHA InfoCard | 100.004.020 |
| KEGG | |
| MeSH | Purine |
PubChem CID
|
|
| UNII | |
CompTox Dashboard (EPA)
|
|
| |
| |
| Properties | |
| C5H4N4 | |
| Molar mass | 120.115 g·mol−1 |
| Melting point | 214 °C (417 °F; 487 K) |
| 500 g/L (RT) | |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
| |
Purine is a heterocyclic aromatic organic compound that consists of two rings (pyrimidine and imidazole) fused together. It is water-soluble. Purine also gives its name to the wider class of molecules, purines, which include substituted purines and their tautomers. They are the most widely occurring nitrogen-containing heterocycles in nature.[1]
Dietary sources
[edit]Purines are found in high concentration in meat and meat products, especially internal organs, such as liver and kidney, and in various seafoods, high-fructose beverages, alcohol, and yeast products.[2][3] Examples of high-purine food sources include anchovies, sardines, liver, beef, kidneys, brains, monkfish, dried mackerel, and shrimp.[3]
Foods particularly rich in hypoxanthine, adenine, and guanine lead to higher blood levels of uric acid.[3] Foods having more than 200 mg of hypoxanthine per 100 g, particularly animal and fish meats containing hypoxanthine as more than 50% of total purines, are more likely to increase uric acid levels.[3] Some vegetables, such as cauliflower, spinach, and peas, have considerable levels of purines, but do not contribute to elevated uric acid levels, possibily due to digestion and bioavailability factors.[3]
Dairy products, soy foods, cereals, beans, mushrooms, and coffee are low-purine foods, characterized specifically by low levels of adenine and guanine comprising more than 60% of purines.[3] A low-purine dietary plan that may reduce the risk of hyperuricemia and gout includes eggs, dairy products, fruits, vegetables, legumes, mushrooms, and soy products.[2][3][4]
Biochemistry
[edit]Purines and pyrimidines make up the two groups of nitrogenous bases, including the two groups of nucleotide bases. The purine bases are guanine (G) and adenine (A) which form corresponding nucleosides-deoxyribonucleosides (deoxyguanosine and deoxyadenosine) with deoxyribose moiety and ribonucleosides (guanosine, adenosine) with ribose moiety. These nucleosides with phosphoric acid form corresponding nucleotides (deoxyguanylate, deoxyadenylate and guanylate, adenylate) which are the building blocks of DNA and RNA, respectively. Purine bases also play an essential role in many metabolic and signalling processes within the compounds guanosine monophosphate (GMP) and adenosine monophosphate (AMP).
In order to perform these essential cellular processes, both purines and pyrimidines are needed by the cell, and in similar quantities. Both purine and pyrimidine are self-inhibiting and activating. When purines are formed, they inhibit the enzymes required for more purine formation. This self-inhibition occurs as they also activate the enzymes needed for pyrimidine formation. Pyrimidine simultaneously self-inhibits and activates purine in a similar manner. Because of this, there is nearly an equal amount of both substances in the cell at all times.[5]
Properties
[edit]Purine is both a very weak acid (pKa 8.93) and an even weaker base (pKa 2.39).[6]
Purine is aromatic, having four tautomers each with a hydrogen bonded to a different one of the four nitrogen atoms. These are identified as 1-H, 3-H, 7-H, and 9-H (see image of numbered ring). The common crystalline form favours the 7-H tautomer, while in polar solvents both the 9-H and 7-H tautomers predominate.[7] Substituents to the rings and interactions with other molecules can shift the equilibrium of these tautomers.[8]
Notable purines
[edit]There are many naturally occurring purines. They include the nucleotide bases adenine and guanine. In DNA, these bases form hydrogen bonds with their complementary pyrimidines, thymine and cytosine, respectively. This is called complementary base pairing. In RNA, the complement of adenine is uracil instead of thymine.
Other notable purines are hypoxanthine, xanthine, theophylline, theobromine, caffeine, uric acid and isoguanine.
Functions
[edit]Aside from the crucial roles of purines (adenine and guanine) in DNA and RNA, purines are also significant components in a number of other important biomolecules, such as ATP, GTP, cyclic AMP, NADH, and coenzyme A. Purine (1) itself, has not been found in nature, but it can be produced by organic synthesis.
They may also function directly as neurotransmitters, acting upon purinergic receptors. Adenosine activates adenosine receptors.
History
[edit]The word purine (pure urine)[9] was coined by the German chemist Emil Fischer in 1884.[10][11] He synthesized it for the first time in 1898.[11] The starting material for the reaction sequence was uric acid (8), which had been isolated from kidney stones by Carl Wilhelm Scheele in 1776.[12] Uric acid was reacted with PCl5 to give 2,6,8-trichloropurine, which was converted with HI and PH4I to give 2,6-diiodopurine. The product was reduced to purine using zinc dust.
Metabolism
[edit]Many organisms have metabolic pathways to synthesize and break down purines.
Purines are biologically synthesized as nucleosides (bases attached to ribose).
Accumulation of modified purine nucleotides is defective to various cellular processes, especially those involving DNA and RNA. To be viable, organisms possess a number of deoxypurine phosphohydrolases, which hydrolyze these purine derivatives removing them from the active NTP and dNTP pools. Deamination of purine bases can result in accumulation of such nucleotides as ITP, dITP, XTP and dXTP.[13]
Defects in enzymes that control purine production and breakdown can severely alter a cell's DNA sequences, which may explain why people who carry certain genetic variants of purine metabolic enzymes have a higher risk for some types of cancer.
Purine biosynthesis in the three domains of life
[edit]Organisms in all three domains of life, eukaryotes, bacteria and archaea, are able to carry out de novo biosynthesis of purines. This ability reflects the essentiality of purines for life. The biochemical pathway of synthesis is very similar in eukaryotes and bacterial species, but is more variable among archaeal species.[14] A nearly complete, or complete, set of genes required for purine biosynthesis was determined to be present in 58 of the 65 archaeal species studied.[14] However, also identified were seven archaeal species with entirely, or nearly entirely, absent purine encoding genes. Apparently the archaeal species unable to synthesize purines are able to acquire exogenous purines for growth.,[14] and are thus analogous to purine mutants of eukaryotes, e.g. purine mutants of the Ascomycete fungus Neurospora crassa,[15] that also require exogenous purines for growth.
Laboratory synthesis
[edit]In addition to in vivo synthesis of purines in purine metabolism, purine can also be synthesized artificially.
Purine is obtained in good yield when formamide is heated in an open vessel at 170 °C for 28 hours.[16]
This reaction and others like it have been discussed in the context of the origin of life.[17]
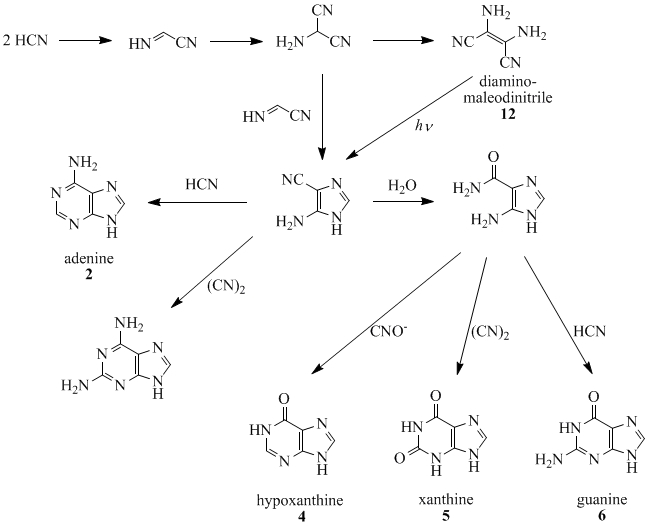
Oro and Kamat (1961) and Orgel co-workers (1966, 1967) have shown that four molecules of HCN tetramerize to form diaminomaleodinitrile (12), which can be converted into almost all naturally occurring purines.[18][19][20][21][22] For example, five molecules of HCN condense in an exothermic reaction to make adenine, especially in the presence of ammonia.
The Traube purine synthesis (1900) is a classic reaction (named after Wilhelm Traube) between an amine-substituted pyrimidine and formic acid.[23]
Prebiotic synthesis of purine ribonucleosides
[edit]In order to understand how life arose, knowledge is required of the chemical pathways that permit formation of the key building blocks of life under plausible prebiotic conditions. Nam et al. (2018)[24] demonstrated the direct condensation of purine and pyrimidine nucleobases with ribose to give ribonucleosides in aqueous microdroplets, a key step leading to RNA formation. Also, a plausible prebiotic process for synthesizing purine ribonucleosides was presented by Becker et al. in 2016.[25]
See also
[edit]References
[edit]- ^ Rosemeyer H (March 2004). "The chemodiversity of purine as a constituent of natural products". Chemistry & Biodiversity. 1 (3): 361–401. doi:10.1002/cbdv.200490033. PMID 17191854. S2CID 12416667.
- ^ a b Li R, Yu K, Li C (2018). "Dietary factors and risk of gout and hyperuricemia: a meta-analysis and systematic review" (PDF). Asia Pacific Journal of Clinical Nutrition. 27 (6): 1344–1356. doi:10.6133/apjcn.201811_27(6).0022. PMID 30485934.
- ^ a b c d e f g Kaneko K, Aoyagi Y, Fukuuchi T, et al. (2014). "Total purine and purine base content of common foodstuffs for facilitating nutritional therapy for gout and hyperuricemia". Biological & Pharmaceutical Bulletin. 37 (5): 709–21. doi:10.1248/bpb.b13-00967. PMID 24553148.
- ^ "Gout diet: What's allowed, what's not". Mayo Clinic. 2 April 2025. Retrieved 13 April 2025.
- ^ Guyton AC (2006). Textbook of Medical Physiology. Elsevier. p. 37. ISBN 978-0-7216-0240-0.
- ^ Seela F, et al. (2014). "Hetarenes III (Six-Membered Rings and Larger Hetero-Rings with Maximum Unsaturation) — Part 2b". In Schaumann E (ed.). Houben-Weyl Methods of Organic Chemistry. Vol. E 9b/2 (4th Supplement ed.). Thieme. p. 310. ISBN 978-3-13-181504-0. Archived from the original on 17 February 2022. Retrieved 15 May 2020.
- ^ Raczyńska ED, Gal JF, Maria PC, et al. (April 2020). "Purine tautomeric preferences and bond-length alternation in relation with protonation-deprotonation and alkali metal cationization". Journal of Molecular Modeling. 26 (5): 93. doi:10.1007/s00894-020-4343-6. PMC 7256107. PMID 32248379.
- ^ Stasyuk OA, Szatyłowicz H, Krygowski TM (April 2012). "Effect of the H-bonding on aromaticity of purine tautomers". The Journal of Organic Chemistry. 77 (8): 4035–45. doi:10.1021/jo300406r. PMID 22448684.
- ^ McGuigan H (1921). An Introduction To Chemical Pharmacology. P. Blakiston's Sons & Co. p. 283. Archived from the original on 16 April 2020. Retrieved 18 July 2012.
- ^ Fischer E (1884). "Ueber die Harnsäure. I." [On uric acid. I.] (PDF). Berichte der Deutschen Chemischen Gesellschaft. 17: 328–338. doi:10.1002/cber.18840170196. Retrieved 20 April 2016.

From p. 329 Archived 2022-02-17 at the Wayback Machine: "Um eine rationelle Nomenklatur der so entstehenden zahlreichen Substanzen zu ermöglichen, betrachte ich dieselben als Abkömmlinge der noch unbekannten Wasserstoffverbindung CH3.C5N4H3 and nenne die letztere Methylpurin." (In order to make possible a rational nomenclature for the numerous existing substances, I regarded them as derivatives of a still unknown hydrogen compound, CH3.C5N4H3, and call the latter "methylpurine".) - ^ a b Fischer E (1898). "Ueber das Purin und seine Methylderivate" [On purine and its methyl derivatives] (PDF). Berichte der Deutschen Chemischen Gesellschaft. 31 (3): 2550–74. doi:10.1002/cber.18980310304. Retrieved 20 April 2016.

From p. 2550 Archived 2020-10-18 at the Wayback Machine: "…hielt ich es für zweckmäßig, alle diese Produkte ebenso wie die Harnsäure als Derivate der sauerstofffreien Verbindung C5H4N4 zu betrachten, und wählte für diese den Namen Purin, welcher aus den Wörtern purum und uricum kombiniert war." (…I regarded it as expedient to consider all of these products, just like uric acid, as derivatives of the oxygen-free compound C5H4N4, and chose for them the name "purine", which was formed from the [Latin] words purum and uricum.) - ^ Scheele CW (1776). "Examen chemicum calculi urinari" [A chemical examination of kidney stones]. Opuscula. 2: 73.
- ^ Davies O, Mendes P, Smallbone K, et al. (April 2012). "Characterisation of multiple substrate-specific (d)ITP/(d)XTPase and modelling of deaminated purine nucleotide metabolism". BMB Reports. 45 (4): 259–264. doi:10.5483/BMBRep.2012.45.4.259. PMID 22531138.
- ^ a b c Brown AM, Hoopes SL, White RH, et al. (2011). "Purine biosynthesis in archaea: Variations on a theme". Biology Direct. 6: 63. doi:10.1186/1745-6150-6-63. PMC 3261824. PMID 22168471.
- ^ Bernstein H (1961). "Imidazole Compounds Accumulated by Purine Mutants of Neurospora crassa". Journal of General Microbiology. 25: 41–46. doi:10.1099/00221287-25-1-41.
- ^ Yamada H, Okamoto T (1972). "A One-step Synthesis of Purine Ring from Formamide". Chemical & Pharmaceutical Bulletin. 20 (3): 623. doi:10.1248/cpb.20.623. Archived from the original on 16 May 2016.
- ^ Saladino R, Crestini C, Ciciriello F, et al. (December 2006). "About a formamide-based origin of informational polymers: syntheses of nucleobases and favourable thermodynamic niches for early polymers". Origins of Life and Evolution of the Biosphere. 36 (5–6): 523–531. Bibcode:2006OLEB...36..523S. doi:10.1007/s11084-006-9053-2. PMID 17136429. S2CID 36278915.
- ^ Sanchez RA, Ferris JP, Orgel LE (December 1967). "Studies in prebiotic synthesis. II. Synthesis of purine precursors and amino acids from aqueous hydrogen cyanide". Journal of Molecular Biology. 30 (2): 223–253. doi:10.1016/S0022-2836(67)80037-8. PMID 4297187.
- ^ Ferris JP, Orgel LE (March 1966). "An Unusual Photochemical Rearrangement in the Synthesis of Adenine from Hydrogen Cyanide". Journal of the American Chemical Society. 88 (5): 1074. doi:10.1021/ja00957a050.
- ^ Ferris JP, Kuder JE, Catalano AW (November 1969). "Photochemical reactions and the chemical evolution of purines and nicotinamide derivatives". Science. 166 (3906): 765–6. Bibcode:1969Sci...166..765F. doi:10.1126/science.166.3906.765. PMID 4241847. S2CID 695243.
- ^ Oro J, Kamat SS (April 1961). "Amino-acid synthesis from hydrogen cyanide under possible primitive earth conditions". Nature. 190 (4774): 442–3. Bibcode:1961Natur.190..442O. doi:10.1038/190442a0. PMID 13731262. S2CID 4219284.
- ^ Bauer W (1985). Houben-Weyl Methods of Organic Chemistry. Vol. E 5 (4th Supplement ed.). Thieme Georg Verlag. p. 1547. ISBN 978-3-13-181154-7.
- ^ Hassner A, Stumer C (2002). Organic Syntheses Based on Name Reactions (2nd ed.). Elsevier. ISBN 0-08-043259-X.
- ^ Nam I, Nam HG, Zare RN (January 2018). "Abiotic synthesis of purine and pyrimidine ribonucleosides in aqueous microdroplets". Proc Natl Acad Sci U S A. 115 (1): 36–40. doi:10.1073/pnas.1718559115. PMC 5776833. PMID 29255025.
- ^ Becker S, Thoma I, Deutsch A, et al. (May 2016). "A high-yielding, strictly regioselective prebiotic purine nucleoside formation pathway". Science. 352 (6287): 833–6. doi:10.1126/science.aad2808. PMID 27174989.
Purine
View on GrokipediaStructure and Properties
Molecular Structure
Purine is a heterocyclic aromatic organic compound consisting of a six-membered pyrimidine ring fused to a five-membered imidazole ring, forming a bicyclic structure that serves as the core scaffold for various nucleobases.[6][7] This fused ring system combines the nitrogen-rich frameworks of pyrimidine (a diazine with nitrogens at positions 1 and 3) and imidazole (a five-membered ring with nitrogens at positions 1 and 3), resulting in a planar, conjugated system with four nitrogen atoms contributing to its aromaticity.[8] The molecular formula of purine is C₅H₄N₄, with a standard numbering system that assigns positions 1, 3, 7, and 9 to the nitrogen atoms and positions 2, 4, 5, 6, and 8 to the carbon atoms, starting from the pyrimidine ring and proceeding through the fusion points at C4-C5 and N7-C8.[9][10] In the predominant 9H-tautomer, the hydrogen is attached to N9, and the structure includes alternating double bonds, such as C2=N3 in the pyrimidine ring and C4=C5 at the fusion site, along with delocalized electrons across both rings to maintain aromatic stability.[8] Purine exists in multiple tautomeric forms due to the mobility of the hydrogen atom among its four nitrogen sites, but the primary tautomers are the 7H and 9H variants, with the equilibrium in solution strongly favoring the 9H form by a significant margin as determined through infrared matrix isolation and ab initio calculations.[11] This preference arises from the energetic stability of the 9H configuration, which better accommodates the aromatic electron distribution in the imidazole ring.[11]Physical Properties
Purine appears as a white to off-white powder under standard conditions.[12] Its molecular formula is C₅H₄N₄, corresponding to a molecular weight of 120.11 g/mol.[9] The compound melts at approximately 214–217 °C, at which point it begins to decompose without a distinct liquid phase.[12] Purine does not have a defined boiling point, as it sublimes at elevated temperatures above its melting point.[13] In terms of solubility, purine is highly soluble in water, with a reported value of 400 g/L at 20 °C; solubility increases further in hot water. It is also soluble in ethanol, toluene, acetone, and hot ethyl acetate, and it forms salts that enhance solubility in dilute acids and bases.[13][14] Spectroscopically, purine exhibits characteristic UV absorption maxima at around 220 nm and 263 nm in neutral aqueous solution, which are attributable to its conjugated heterocyclic ring system and are commonly used for its quantitative detection in biochemical assays.[15]Chemical Properties
Purine displays amphoteric behavior, acting as both a weak base and a weak acid due to the presence of nitrogen atoms in its fused ring system. The pKa value for protonation at the N1 position, reflecting its basicity, is approximately 2.4, while the pKa for deprotonation at the N9-H, indicating its acidity, is approximately 9.8.[16][17] These values position purine in a neutral form under physiological conditions, with protonation favored in strongly acidic media and deprotonation in basic environments. The stability of purine is significantly enhanced by its aromatic character, arising from a delocalized π-electron system across the imidazole and pyrimidine rings, which follows Hückel's rule with 10 π electrons. This resonance delocalization contributes to the molecule's resistance to thermal decomposition and confers planarity to the ring system, minimizing strain and promoting overall thermodynamic stability.[8] In terms of reactivity, purine undergoes electrophilic substitution primarily at the C8 and C2 positions of the imidazole and pyrimidine rings, respectively, due to the relative electron density at these sites facilitated by the electron-rich aromatic framework. Conversely, nucleophilic aromatic substitution occurs at the C6 and C2 positions, particularly when activated by good leaving groups such as halogens, enabling displacement under milder conditions at C6 compared to C2.[18][19] Purine exhibits a moderate oxidation potential, rendering it susceptible to one-electron oxidation under mild conditions to form radical cations or derivatives like 8-hydroxypurine, often initiated by reactive oxygen species. This reactivity underscores its role in oxidative stress pathways but also highlights vulnerability to environmental oxidants. Regarding stability in aqueous media, purine remains resistant to hydrolysis at neutral pH, maintaining structural integrity over extended periods, though it shows sensitivity to strong oxidizing agents that can disrupt the ring system.[20]Biological Functions
Role in Nucleic Acids
Purines play a central role in the structure and function of nucleic acids, serving as two of the four nucleobases in both DNA and RNA. Adenine, chemically known as 6-aminopurine, and guanine, known as 2-amino-6-oxopurine, are the purine components that integrate into these biopolymers.[21][22] These bases attach to a sugar moiety—deoxyribose in DNA or ribose in RNA—to form nucleosides such as deoxyadenosine, adenosine, deoxyguanosine, and guanosine. Further phosphorylation of these nucleosides at the 5' position of the sugar yields nucleotides, including deoxyadenosine monophosphate (dAMP), adenosine monophosphate (AMP), deoxyguanosine monophosphate (dGMP), and guanosine monophosphate (GMP), which are the monomeric units polymerized into DNA and RNA strands.[23] The specific base-pairing rules governed by hydrogen bonds ensure the fidelity of genetic information storage and transfer in nucleic acids. In DNA, adenine pairs with thymine via two hydrogen bonds, while guanine pairs with cytosine via three hydrogen bonds, contributing to the stability of the double helix.[24] In RNA, adenine pairs with uracil through two hydrogen bonds, and guanine continues to pair with cytosine using three, facilitating processes like mRNA-tRNA interactions during translation.[23] This complementary pairing adheres to Chargaff's rules, which dictate that in double-stranded DNA, the proportion of purine bases (adenine plus guanine) equals that of pyrimidine bases (thymine plus cytosine), resulting in approximately 50% of the bases being purines.[25] Beyond hydrogen bonding, purines contribute to the structural integrity of the nucleic acid double helix through base-stacking interactions. Adjacent bases along the helix axis engage in π-π stacking, where the planar aromatic rings of purines like adenine and guanine overlap with neighboring bases, providing hydrophobic stabilization and resisting unwinding forces.[26] These stacking interactions, particularly involving purine-purine pairs, enhance the overall rigidity and thermal stability of the helical structure, complementing the specificity of base pairing to maintain the genetic blueprint.[27]Roles in Metabolism and Signaling
Purines play essential roles in cellular energy metabolism through their incorporation into nucleoside triphosphates such as adenosine triphosphate (ATP) and guanosine triphosphate (GTP), which serve as universal energy currencies. The high-energy phosphoanhydride bonds in these molecules enable the storage and transfer of energy for endergonic processes, including biosynthesis, transport, and mechanical work. For instance, the hydrolysis of ATP to adenosine diphosphate (ADP) and inorganic phosphate (P_i) releases energy via the reaction ATP → ADP + P_i, with a standard free energy change (\Delta G^{\circ'}) of approximately -30.5 kJ/mol under physiological conditions.[28] Similarly, GTP hydrolysis powers specific reactions, such as protein synthesis during translation, highlighting the complementary functions of these purine-based triphosphates in maintaining cellular energy homeostasis.[29] Beyond energy transfer, purines contribute to redox metabolism as components of key coenzymes. Nicotinamide adenine dinucleotide (NAD^+), which contains the purine base adenine, functions as an electron carrier in catabolic pathways like glycolysis, the tricarboxylic acid cycle, and fatty acid oxidation, facilitating hydride transfer in two-electron redox reactions.[30] Flavin adenine dinucleotide (FAD), also featuring adenine, participates in flavoprotein-mediated oxidations, such as those in the electron transport chain and amino acid metabolism, where it accepts electrons to form FADH_2.[31] These coenzymes link purine structures to the regulation of oxidative processes essential for ATP production and cellular respiration. In cellular signaling, purine derivatives act as critical second messengers that amplify extracellular signals. Cyclic adenosine monophosphate (cAMP), derived from ATP, mediates responses to hormones like glucagon and epinephrine by activating protein kinase A, which phosphorylates targets to influence glycogenolysis, gene expression, and ion channel activity.[32] Cyclic guanosine monophosphate (cGMP), formed from GTP, regulates pathways involving nitric oxide and atrial natriuretic peptide, modulating smooth muscle relaxation, phototransduction in vision, and vascular homeostasis through activation of protein kinase G.[33] These cyclic nucleotides enable rapid, localized signal transduction, distinguishing purines' dynamic regulatory roles from their structural functions in nucleic acids, where they primarily facilitate base pairing. Purines also function in protective and salvage mechanisms, with uric acid serving as a potent antioxidant in species like humans that lack uricase enzyme activity. Uric acid neutralizes reactive oxygen species, mitigating oxidative stress in conditions such as inflammation and neurodegeneration, thereby contributing to evolutionary adaptations in metabolic resilience. Additionally, hypoxanthine, a purine intermediate, is central to the salvage pathway, where it is recycled by hypoxanthine-guanine phosphoribosyltransferase to form nucleotides, conserving energy and preventing wasteful de novo synthesis.[34] The involvement of purines in these processes underscores their evolutionary conservation as hubs in core metabolic networks. Purine metabolism genes have undergone selective pressures across mammals, enhancing oxidative stress adaptation and integrating into universal pathways like energy production and signaling, which trace back to early cellular life forms.[35] This conservation reflects purines' foundational role in linking catabolism, redox balance, and environmental responsiveness across diverse organisms.[36]Metabolism
Biosynthesis
Purine nucleotides are synthesized endogenously through two primary pathways: the de novo biosynthesis pathway, which constructs the purine ring from simple precursors, and the salvage pathway, which recycles free purine bases.[37] The de novo pathway is a 10-step process that begins with phosphoribosyl pyrophosphate (PRPP) and culminates in the formation of inosine monophosphate (IMP), the first purine nucleotide.[38] This pathway requires six enzymes in eukaryotes, including glutamine-PRPP amidotransferase (also known as PRPP amidotransferase), which catalyzes the committed first step by transferring an amide group from glutamine to PRPP, forming phosphoribosylamine (PRA); glycinamide ribonucleotide (GAR) transformylase, which adds a formyl group in the third step; and 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) transformylase, which performs the final formylation in the 10th step.[39] The process consumes six high-energy phosphate bonds equivalent to ATP molecules per IMP produced, highlighting its metabolic cost.[37] The salvage pathway provides a more energy-efficient alternative by reutilizing purine bases derived from dietary sources or nucleotide degradation. Key enzymes include hypoxanthine-guanine phosphoribosyltransferase (HGPRT), which catalyzes the conversion of hypoxanthine to IMP and guanine to guanosine monophosphate (GMP) using PRPP, and adenine phosphoribosyltransferase (APRT), which converts adenine to adenosine monophosphate (AMP).[40] These reactions conserve PRPP and bypass the energy-intensive de novo assembly, making salvage particularly important in tissues with high nucleotide turnover, such as the brain.[40] Variations in purine biosynthesis exist across biological domains. In bacteria, the pathway is often organized into the Pur operon, a coordinated gene cluster that facilitates efficient regulation and expression under varying nutrient conditions.[39] Eukaryotes perform de novo synthesis entirely in the cytosol, with enzymes forming dynamic complexes known as purinosomes to enhance efficiency.[41] In archaea, the pathway exhibits greater variability, including distinct amidotransferases for the initial step and alternative enzymes for certain transformations, such as non-homologous versions of AIR carboxylase and SAICAR synthetase, reflecting adaptations to extreme environments.[42] Regulation of purine biosynthesis primarily occurs through feedback inhibition at early steps to prevent overproduction. The rate-limiting enzyme, PRPP amidotransferase, is allosterically inhibited by binding of AMP and GMP to separate regulatory sites, with synergistic effects when both are present; IMP also exerts milder inhibition.[3] This end-product inhibition balances nucleotide pools according to cellular needs. IMP serves as a branch point precursor, being converted to AMP via adenylosuccinate synthetase and lyase or to GMP via IMP dehydrogenase and GMP synthetase.[38]Catabolism and Uric Acid Formation
Purine catabolism involves the sequential degradation of purine nucleotides to nucleosides and then to free bases, ultimately leading to uric acid as the primary end product in humans. This process begins with the dephosphorylation of nucleotides such as adenosine monophosphate (AMP) to adenosine by 5'-nucleotidase, followed by the deamination of adenosine to inosine by adenosine deaminase (ADA). Inosine is then converted to hypoxanthine via phosphorolysis catalyzed by purine nucleoside phosphorylase (PNP). Similarly, guanosine monophosphate (GMP) follows a parallel path through guanosine to guanine, which is deaminated to xanthine by guanine deaminase.[43][44][1] Hypoxanthine is oxidized to xanthine by xanthine oxidase (XO), and xanthine is further oxidized to uric acid by the same enzyme, marking the terminal steps of purine breakdown. XO catalyzes these reactions using molecular oxygen as the electron acceptor, producing hydrogen peroxide (H₂O₂) as a byproduct in each step, with two molecules of H₂O₂ generated overall during the conversion of hypoxanthine to uric acid. Some catabolic intermediates, such as hypoxanthine and guanine, can be briefly recycled into nucleotides via salvage pathways.[45][46][43] Species exhibit significant variations in purine catabolism due to differences in the enzyme urate oxidase (uricase). In humans and other hominoid primates, functional uricase is absent, resulting in uric acid as the end product, which is less soluble than further metabolites. Birds and reptiles also excrete uric acid as their primary nitrogenous waste, aiding in water conservation. In contrast, most other mammals possess active uricase, which oxidizes uric acid to the more soluble allantoin for excretion.[47][36][48] In humans, approximately 70% of daily uric acid production arises from endogenous turnover of nucleic acids and nucleotides, with the remainder from dietary sources. Uric acid is primarily excreted via the kidneys, accounting for about 70% of total elimination, while the intestines handle the rest through bacterial degradation. Imbalances in this catabolic pathway can lead to hyperuricemia, defined by serum uric acid levels exceeding 6.8 mg/dL—the saturation threshold at physiological pH and temperature—which promotes monosodium urate crystal formation and gout.[49][50][51]Sources and Synthesis
Dietary Sources
Purines and their derivatives, such as adenine and guanine, are naturally occurring compounds found in various foods, particularly those of animal origin, though plant-based sources also contribute in smaller amounts.[52] High-purine foods are often energy-dense and include organ meats and certain seafood, which can significantly influence uric acid levels upon metabolism.[53] Organ meats represent some of the richest dietary sources of purines, with values typically ranging from 100 to 400 mg per 100 g. For instance, beef liver contains approximately 220–231 mg/100 g, while beef kidney has about 112–174 mg/100 g.[54][55] Seafood, another high-purine category, often exceeds 200 mg/100 g; anchovies provide around 239–321 mg/100 g, and sardines are similarly elevated at 210–480 mg/100 g depending on preparation.[54] Yeast extracts, used in products like spreads and broths, can contain even higher levels, up to 600–1000 mg/100 g, making them potent sources.[52] Moderate-purine foods include red meats and poultry, generally providing 50–150 mg/100 g. Beef cuts range from 77 to 123 mg/100 g, and chicken is comparable at 50–140 mg/100 g.[56] Legumes such as beans and lentils fall in a similar range of 50–100 mg/100 g, with dry black mung beans reaching up to 222 mg/100 g in some varieties, though most are lower. Low-purine options, suitable for restricting intake, encompass dairy products, most vegetables, and grains, all typically under 50 mg/100 g. Milk and cheeses contain less than 10 mg/100 g, while vegetables like spinach or asparagus may reach 20–50 mg/100 g but are still considered minimal contributors. Grains such as rice or bread provide negligible amounts, often below 20 mg/100 g.[52] In typical Western diets, daily purine intake averages 600–1000 mg, primarily from meat, seafood, and processed foods.[57] These dietary purines are absorbed mainly in the small intestine as nucleosides via concentrative nucleoside transporters, before being metabolized to uric acid in the liver and other tissues.[58] This exogenous input supplements the endogenous purine pool, with catabolism ultimately yielding uric acid as the primary end product.[52]| Category | Examples | Purine Content (mg/100 g) |
|---|---|---|
| High | Beef liver, anchovies, yeast extracts | 200–1000 |
| Moderate | Beef, chicken, lentils | 50–150 |
| Low | Milk, spinach, rice | <50 |