Recent from talks
Nothing was collected or created yet.
HLA-DR
View on Wikipedia| MHC class II, DR | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (heterodimer) | |||||||||||||||||||
Illustration of DR with bound ligand (yellow) | |||||||||||||||||||
| Protein type | cell surface receptor | ||||||||||||||||||
| Function | Immune recognition and antigen presentation | ||||||||||||||||||
| |||||||||||||||||||
HLA-DR is an MHC class II cell surface receptor encoded by the human leukocyte antigen complex on chromosome 6 region 6p21.31. The complex of HLA-DR (Human Leukocyte Antigen – DR isotype) and peptide, generally between 9 and 30 amino acids in length, constitutes a ligand for the T-cell receptor (TCR). HLA (human leukocyte antigens) were originally defined as cell surface antigens that mediate graft-versus-host disease. Identification of these antigens has led to greater success and longevity in organ transplant.
Antigens most responsible for graft loss are HLA-DR (first six months), HLA-B (first two years), and HLA-A (long-term survival).[1] Good matching of these antigens between host and donor is most critical for achieving graft survival.
HLA-DR is also involved in several autoimmune conditions, disease susceptibility and disease resistance. It is also closely linked to HLA-DQ and this linkage often makes it difficult to resolve the more causative factor in disease.
HLA-DR molecules are upregulated in response to signalling. In the instance of an infection, the peptide (such as the staphylococcal enterotoxin I peptide) is bound into a DR molecule and presented to a few of a great many T-cell receptors found on T-helper cells. These cells then bind to antigens on the surface of B-cells stimulating B-cell proliferation.
Function
[edit]
The primary function of HLA-DR is to present peptide antigens, potentially foreign in origin, to the immune system for the purpose of eliciting or suppressing T-(helper)-cell responses that eventually lead to the production of antibodies against the same peptide antigen. Antigen-presenting cells (macrophages, B-cells and dendritic cells) are the cells in which DR are typically found. Increased abundance of DR 'antigen' on the cell surface is often in response to stimulation, and, therefore, DR is also a marker for immune stimulation.
Structure
[edit]HLA-DR is an αβ heterodimer, cell surface receptor, each subunit of which contains two extracellular domains, a membrane-spanning domain and a cytoplasmic tail. Both α and β chains are anchored in the membrane. The N-terminal domain of the mature protein forms an alpha-helix that constitutes the exposed part of the binding groove, the C-terminal cytoplasmic region interact with the other chain forming a beta-sheet under the binding groove spanning to the cell membrane. The majority of the peptide contact positions are in the first 80 residues of each chain.
Genetics
[edit]The genetics of HLA-DR is complex. HLA-DR is encoded by several loci and several 'genes' of different function at each locus. The DR α-chain is encoded by the HLA-DRA locus. Unlike the other DR loci, functional variation in mature DRA gene products is absent. (Note: see table Number of Variant Alleles HLA-DR Loci) This reduces the potential functional combinations from ~1400 to ~400 ([table is not exact because new alleles are continually being added; not all new alleles are functional variants of the mature subunits]).
| DR | DR-DQ | DR | DQ | Freq | |||
|---|---|---|---|---|---|---|---|
| Serotype | haplotype | B1 | A1 | B1 | %[2] | ||
| DR1 | DR1-DQ5 | 01:01 | 01:01 | 05:01 | 9. | 1 | |
| 01:02 | 01:01 | 05:01 | 1. | 4 | |||
| 01:03 | 01:01 | 05:01 | 0. | 5 | |||
| DR3 | DR3-DQ2 | 03:01 | 05:01 | 02:01 | 13. | 1 | |
| DR4 | DR4-DQ7 | 04:01 | 0300 | 03:01 | 5. | 4 | |
| 04:07 | 0300 | 03:01 | 0. | 9 | |||
| DR4-DQ8 | 04:01 | 0300 | 03:02 | 5. | 0 | ||
| 04:02 | 0300 | 03:02 | 1. | 0 | |||
| 04:03 | 0300 | 03:02 | 0. | 4 | |||
| 04:04 | 0300 | 03:02 | 3. | 9 | |||
| 04:05 | 0300 | 03:02 | 0. | 3 | |||
| DR7 | DR7-DQ2 | 07:01 | 02:01 | 02:02 | 11. | 1 | |
| DR7-DQ9 | 07:01 | 02:01 | 03:03 | 3. | 7 | ||
| DR8 | DR8-DQ4 | 08:01 | 04:01 | 04:02 | 2. | 2 | |
| DR8-DQ7 | 08:03 | 06:01 | 03:01 | 0. | 1 | ||
| DR9 | DR9-DQ9 | 09:01 | 0300 | 03:03 | 0. | 8 | |
| DR10 | DR10-DQ5 | 10:01 | 01:04 | 05:01 | 0. | 7 | |
| DR11 | DR11-DQ7 | 11:01 | 05:05 | 03:01 | 5. | 6 | |
| 11:03 | 05:05 | 03:01 | 0. | 3 | |||
| 11:04 | 05:05 | 03:01 | 2. | 7 | |||
| DR12 | DR12-DQ7 | 12:01 | 05:05 | 03:01 | 1. | 1 | |
| DR13 | DR13-DQ6 | 13:01 | 01:03 | 06:03 | 5. | 6 | |
| 13:02 | 01:02 | 06:04 | 3. | 4 | |||
| 13:02 | 01:02 | 06:09 | 0. | 7 | |||
| DR13-DQ7 | 13:03 | 05:05 | 03:01 | 0. | 7 | ||
| DR14 | DR14-DQ5 | 14:01 | 01:04 | 05:03 | 2. | 0 | |
| DR15 | DR15-DQ6 | 15:01 | 01:02 | 06:02 | 14. | 2 | |
| 15:02 | 01:03 | 06:01 | 0. | 7 | |||
| DR16 | DR16-DQ5 | 16:01 | 01:02 | 05:02 | 1. | 0 | |
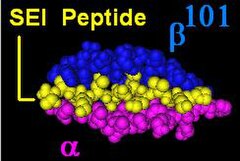
The DR β-chain[3] is encoded by 4 loci, however no more than 3 functional loci are present in a single individual, and no more than two on a single chromosome. Sometimes an individual may only possess 2 copies of the same locus, DRB1*. The HLA-DRB1 locus is ubiquitous and encodes a very large number of functionally variable gene products (HLA-DR1 to HLA-DR17). The HLA-DRB3 locus encodes the HLA-DR52 specificity, is moderately variable and is variably associated with certain HLA-DRB1 types. The HLA-DRB4 locus encodes the HLA-DR53 specificity, has some variation, and is associated with certain HLA-DRB1 types. The HLA-DRB5 locus encodes the HLA-DR51 specificity, which is typically invariable, and is linked to the HLA-DR2 types.
- linkage (See Table)
- DQA1 and DQB1
- Linkage disequilibrium exists for many DR-DQ types.
- Nomenclature issues. Some older studies may refer to DR15 or 16 as DR2 and DQ5 and DQ6 as DQ1 therefore a haplotype DR2-DQ1 is usually referring to DR15-DQ6 but could be referring to DR16-DQ5. DR5 is used to refer to DR11 and DR12, in which case DQ3 might be used. In these instances DQ3 almost always can be interpreted as DQ7, but DR5 is most often DR11 and less frequently DR12. Similar issues exist for DR6 versus DR13 and DR14. DR6-DQ1 can refer to either DR13-DQ6 or less frequently DR14-DQ5, but DR6-DQ3 or DR6-DQ7 generally refers to DR13-DQ7. Even older literature has more confusing designations. By looking at the change of disease association with improved testing we can see how HLA nomenclature has evolved over time.
- DQA1 and DQB1
| HLA-DR | ||||
|---|---|---|---|---|
| HLA | -A1 | -B1 | -B3 to -B51 | Potential |
| Locus | # | # | # | Combinations |
| Alleles[3][4] | 3 | 463 | 74 | 1635 |
| Unique Polypeptide | 2 | 394 | 57 | 902 |
| Contact Variant | 1 | ~300 | ~30 | ~330 |
| 1DRB3, DRB4, DRB5 have variable presence in humans | ||||
Evolution and allele frequencies
[edit]There is a high level of allelic diversity at HLA DRB1, it is second only to HLA-B locus in number of allelic variants. These two loci are highest sequence variation rate within human genome. This means HLA-DRB1 is rapidly evolving, much more rapidly than almost all other protein encoding loci. Much of the variation at HLA DRB1 occurs at peptide contact positions in the binding groove, as a result many of the alleles alter the way the DR binds peptide ligands and changes the repertoire each receptor can bind. This means that most of the changes are functional in nature, and therefore are under selection. In the HLA region, genes are under heterozygous or balancing selection, although certain alleles appear to be under positive or negative selection, either in the past or present
HLA generally evolve through a process of gene conversion, which is a form of short distance or 'abortive' genetic recombination. Functional motifs in genes are exchanged to form new alleles, and frequently new, functionally different DR isoforms. HLA-DR represents an extreme example of this. A survey of X-linked loci reveals that most human loci have undergone fixation within the last 600,000 years, and diploid loci have undergone significant proportion of fixation in that period of time.
The level of deep branching at X-linked loci indicates loci were close to fixation or fixed at the end of the human population bottleneck 100,000 to 150,000 years ago. The HLA-DR locus represents a major exception to this observation.[5] Based on distribution of major groupings in the human population it is possible to assert that more than a dozen major variants survived the population bottleneck. This observation is supported by the concept of a heterozygous selection coefficient operating on the HLA-DR, and at the HLA-DRB1 locus to a greater degree relative to HLA-DQB1 and HLA-DPB1. Most of the HLA alleles currently present in the human population can be explained by gene conversion between these ancient ancestral types,[6] some that persist into the extant population.
Serogroups
[edit]| Serotypes of HLA-DRB1 gene products | ||
| Split antigens | ||
| HLA-DR1 | ||
| HLA-DR2 | HLA-DR15 | HLA-DR16 |
| HLA-DR3 | HLA-DR17 | HLA-DR18 |
| HLA-DR4 | ||
| HLA-DR5 | HLA-DR11 | HLA-DR12 |
| HLA-DR6 | HLA-DR13 | HLA-DR14 |
| HLA-DR7 | ||
| HLA-DR8 | ||
| HLA-DR9 | ||
| HLA-DR10 | ||
The table below provides links to subpages with information about distribution, genetic linkage and disease association for the HLA-DR serogroups.
Interlocus DRB linkage
[edit]DRB1 is linked with other DRB loci in four ways.
| non-DRB1 | linked DRB1 antigens | |||
|---|---|---|---|---|
| antigens | antigens | |||
| None | DR1 | DR8 | DR10 | |
| DR51 | DR2 | DR15 | DR16 | |
| DR52 | DR3 | DR17 | DR18 | |
| DR5 | DR11 | DR12 | ||
| DR6 | DR13 | DR14 | ||
| DR53 | DR4 | DR7 | DR8 | DR9 |
| Class | Disease | Associated DR | 2 | 3 | 4 |
|---|---|---|---|---|---|
| alopecia areata | DR5 | ||||
| anemia | pernicious | DR15 | |||
| antiphospholipid syndrome, primary | DR5 | DR12 | |||
| aneurysm | coronary artery | DR16 | |||
| arteritis | Takayasu's | DR16 | |||
| arthritis, rheumatoid | juvenile | DR4 | DR5 | DR14 | DR15 |
| pauciarticular, juv. | DR8 | ||||
| Still's disease | DR12 | ||||
| iritis w/juv. arthritis | DR12 | ||||
| seropositive | DR1 | DR4 | DR10 | ||
| w/systemic sclerosis | DR1 | ||||
| lyme disease induced | DR4 | ||||
| tiopronin intolerance | DR5 | DR11 | DR12 | ||
| cardiomyopathy | hypertrophic | DR4 | DR17 | ||
| T. cruzi induced | DR4 | DR7 | DR15 | ||
| colitis | Crohn's | DR1 | |||
| ulcerative | DR1 | ||||
| diabetes | juvenile (type 1) | DR3 | DR4 | DR17 | DR18 |
| fatty liver (type 2) | DR8 | ||||
| encephalomyelitis | rabies vaccine-induced | DR17 | |||
| encephalopathy | acute necrotizing | DR52 | |||
| epilepsy | childhood | DR5 | |||
| infantile/spasm | DR17 | ||||
| heart disease | rheumatic | DR16 | |||
| hepatitis | autoimmune | DR2 | DR4 | DR17 | |
| primary biliary cirrhosis | DR2 | DR8 | |||
| chronic type C | DR11 | ||||
| lichen planus | DR1 | DR10 | |||
| lupus, | systemic | DR3 | DR4 | DR52 | |
| hydralazine-induced | DR4 | ||||
| with Sjögren syndrome | DR15 | ||||
| lymphadenopathy | generalized | DR5 | |||
| lymphoma, | mycosis fungoides | DR5 | |||
| melioidosis | DR16 | ||||
| myasthenia | gravis | DR3 | DR6 | DR13 | DR14 |
| penicillamine-induced | DR1 | ||||
| myositis | inflammatory inclusion body | DR17 | DR18 | DR52 | |
| narcolepsy | DR2 | DR12 | |||
| nephritis, | tubulointerstitial | DR1 | |||
| nephropathy | IgA-mediated | DR4 | |||
| polyglandular deficiency syndrome | DR5 | ||||
| pemphigus | foliaceous | DR1 | |||
| vulgaris | DR4 | ||||
| psoriasis | vulgaris | DR1 | DR7 | ||
| papillomatosis, | respiratory | DR1 | |||
| sarcoidosis | non-chronic | DR17 | DR52 | ||
| sclerosis, | multiple | DR2 | DR15 | DR53 | |
| "bout onset" multiple | DR3 | ||||
| systemic | DR4 | DR11 | DR16 | DR52 | |
| vulval lichen | DR12 | ||||
| schizophrenia | DR1 | ||||
| susceptibility | leprosy | DR2 | |||
| tuberculosis | DR2 | ||||
| ragweed Ra6 allergy | DR5 | ||||
| asthma, mite sensitive | DR11 | ||||
| 2ndary infection, AIDS | DR3 | ||||
| aspergillosis | DR15 | ||||
| Kaposi's sarcoma | DR5 | ||||
| thyroid carcinomas | DR8 | DR11 | |||
| ovarian/cervical cancer | DR10 | DR11 | DR15 | ||
| grape induced anaphylaxis | DR11 | ||||
| Chlamydia pneumoniae | DR52 | ||||
| thyroiditis | Hashimoto's | DR3 | DR5 | ||
| Graves' | DR3 | DR17 | DR52 | ||
| uveitis | tubulointerstitial | DR1 | |||
| *references are provided on linked subpages | |||||
References
[edit]- ^ Solomon S, Pitossi F, Rao MS (2015). "Banking on iPSC--is it doable and is it worthwhile". Stem Cell Reviews. 11 (1): 1–10. doi:10.1007/s12015-014-9574-4. PMC 4333229. PMID 25516409.
- ^ Klitz W, Maiers M, Spellman S, Baxter-Lowe LA, Schmeckpeper B, Williams TM, Fernandez-Vina M (2003). "New HLA haplotype frequency reference standards: high-resolution and large sample typing of HLA DR-DQ haplotypes in a sample of European Americans". Tissue Antigens. 62 (4): 296–307. doi:10.1034/j.1399-0039.2003.00103.x. PMID 12974796.
- ^ a b Marsh, S. G.; Albert, E. D.; Bodmer, W. F.; Bontrop, R. E.; Dupont, B.; Erlich, H. A.; Fernández-Viña, M.; Geraghty, D. E.; Holdsworth, R.; Hurley, C. K.; Lau, M.; Lee, K. W.; Mach, B.; Maiers, M.; Mayr, W. R.; Müller, C. R.; Parham, P.; Petersdorf, E. W.; Sasazuki, T.; Strominger, J. L.; Svejgaard, A.; Terasaki, P. I.; Tiercy, J. M.; Trowsdale, J. (2010). "Nomenclature for factors of the HLA system, 2010". Tissue Antigens. 75 (4): 291–455. doi:10.1111/j.1399-0039.2010.01466.x. PMC 2848993. PMID 20356336.
- ^ Robinson J, Waller M, Parham P, de Groot N, Bontrop R, Kennedy L, Stoehr P, Marsh S (2003). "IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex". Nucleic Acids Res. 31 (1): 311–4. doi:10.1093/nar/gkg070. PMC 165517. PMID 12520010.
- ^ Ayala F (1995). "The myth of Eve: molecular biology and human origins" (PDF). Science. 270 (5244): 1930–6. Bibcode:1995Sci...270.1930A. doi:10.1126/science.270.5244.1930. PMID 8533083.
- ^ Parham P, Ohta T (1996). "Population biology of antigen presentation by MHC class I molecules". Science. 272 (5258): 67–74. Bibcode:1996Sci...272...67P. doi:10.1126/science.272.5258.67. PMID 8600539. S2CID 22209086.
Further reading
[edit]- Bénichou S, Benmerah A (2003). "The HIV nef and the Kaposi-sarcoma-associated virus K3/K5 proteins: "parasites"of the endocytosis pathway". Med Sci (Paris). 19 (1): 100–6. doi:10.1051/medsci/2003191100. PMID 12836198.
- Tolstrup M, Ostergaard L, Laursen AL, et al. (2004). "HIV/SIV escape from immune surveillance: focus on Nef". Curr. HIV Res. 2 (2): 141–51. doi:10.2174/1570162043484924. PMID 15078178.
- Anderson JL, Hope TJ (2005). "HIV accessory proteins and surviving the host cell". Current HIV/AIDS Reports. 1 (1): 47–53. doi:10.1007/s11904-004-0007-x. PMID 16091223. S2CID 34731265.
- Li L, Li HS, Pauza CD, et al. (2006). "Roles of HIV-1 auxiliary proteins in viral pathogenesis and host-pathogen interactions". Cell Res. 15 (11–12): 923–34. doi:10.1038/sj.cr.7290370. PMID 16354571.
- Stove V, Verhasselt B (2006). "Modelling thymic HIV-1 Nef effects". Curr. HIV Res. 4 (1): 57–64. doi:10.2174/157016206775197583. PMID 16454711.
- Matsushima GK, Itoh-Lindstrom Y, Ting JP (1992). "Activation of the HLA-DRA gene in primary human T lymphocytes: novel usage of TATA and the X and Y promoter elements". Mol. Cell. Biol. 12 (12): 5610–9. doi:10.1128/MCB.12.12.5610. PMC 360500. PMID 1448091.
- Schaiff WT, Hruska KA, McCourt DW, et al. (1992). "HLA-DR associates with specific stress proteins and is retained in the endoplasmic reticulum in invariant chain negative cells". J. Exp. Med. 176 (3): 657–66. doi:10.1084/jem.176.3.657. PMC 2119345. PMID 1512535.
- Piatier-Tonneau D, Gastinel LN, Amblard F, et al. (1991). "Interaction of CD4 with HLA class II antigens and HIV gp120". Immunogenetics. 34 (2): 121–8. doi:10.1007/BF00211424. PMID 1869305. S2CID 10116507.
- Nong Y, Kandil O, Tobin EH, et al. (1991). "The HIV core protein p24 inhibits interferon-gamma-induced increase of HLA-DR and cytochrome b heavy chain mRNA levels in the human monocyte-like cell line THP1". Cell. Immunol. 132 (1): 10–6. doi:10.1016/0008-8749(91)90002-S. PMID 1905983.
- Rosenstein Y, Burakoff SJ, Herrmann SH (1990). "HIV-gp120 can block CD4-class II MHC-mediated adhesion". J. Immunol. 144 (2): 526–31. doi:10.4049/jimmunol.144.2.526. PMID 1967269. S2CID 23550626.
- Callahan KM, Fort MM, Obah EA, et al. (1990). "Genetic variability in HIV-1 gp120 affects interactions with HLA molecules and T cell receptor". J. Immunol. 144 (9): 3341–6. doi:10.4049/jimmunol.144.9.3341. PMID 1970352. S2CID 23599258.
- Bowman MR, MacFerrin KD, Schreiber SL, Burakoff SJ (1991). "Identification and structural analysis of residues in the V1 region of CD4 involved in interaction with human immunodeficiency virus envelope glycoprotein gp120 and class II major histocompatibility complex molecules". Proc. Natl. Acad. Sci. U.S.A. 87 (22): 9052–6. doi:10.1073/pnas.87.22.9052. PMC 55099. PMID 1978941.
- Koppelman B, Cresswell P (1990). "Rapid nonlysosomal degradation of assembled HLA class II glycoproteins incorporating a mutant DR alpha-chain". J. Immunol. 145 (8): 2730–6. doi:10.4049/jimmunol.145.8.2730. PMID 2212658. S2CID 26256828.
- Clayton LK, Sieh M, Pious DA, Reinherz EL (1989). "Identification of human CD4 residues affecting class II MHC versus HIV-1 gp120 binding". Nature. 339 (6225): 548–51. Bibcode:1989Natur.339..548C. doi:10.1038/339548a0. PMID 2543930. S2CID 4246781.
- Diamond DC, Sleckman BP, Gregory T, et al. (1988). "Inhibition of CD4+ T cell function by the HIV envelope protein, gp120". J. Immunol. 141 (11): 3715–7. doi:10.4049/jimmunol.141.11.3715. PMID 2846691. S2CID 2607172.
- Tjernlund U, Scheynius A, Johansson C, et al. (1989). "T-cell response to purified protein derivative after removal of Langerhans' cells from epidermal cell suspensions containing keratinocytes expressing class II transplantation antigens". Scand. J. Immunol. 28 (6): 667–73. doi:10.1111/j.1365-3083.1988.tb01500.x. PMID 3266023. S2CID 25824282.
- Andrieu JM, Even P, Venet A (1986). "AIDS and related syndromes as a viral-induced autoimmune disease of the immune system: an anti-MHC II disorder. Therapeutic implications". AIDS Research. 2 (3): 163–74. doi:10.1089/aid.1.1986.2.163. PMID 3489470.
- Das HK, Lawrance SK, Weissman SM (1983). "Structure and nucleotide sequence of the heavy chain gene of HLA-DR". Proc. Natl. Acad. Sci. U.S.A. 80 (12): 3543–7. Bibcode:1983PNAS...80.3543D. doi:10.1073/pnas.80.12.3543. PMC 394085. PMID 6304715.
- Schamboeck A, Korman AJ, Kamb A, Strominger JL (1984). "Organization of the transcriptional unit of a human class II histocompatibility antigen: HLA-DR heavy chain". Nucleic Acids Res. 11 (24): 8663–75. doi:10.1093/nar/11.24.8663. PMC 326615. PMID 6324094.
- Das HK, Biro PA, Cohen SN, et al. (1983). "Use of synthetic oligonucleotide probes complementary to genes for human HLA-DR alpha and beta as extension primers for the isolation of 5'-specific genomic clones". Proc. Natl. Acad. Sci. U.S.A. 80 (6): 1531–5. Bibcode:1983PNAS...80.1531D. doi:10.1073/pnas.80.6.1531. PMC 393635. PMID 6403940.
External links
[edit]- HLA-DR+antigens at the U.S. National Library of Medicine Medical Subject Headings (MeSH)