Recent from talks
Contribute something
Nothing was collected or created yet.
DNA sequencing
View on Wikipedia
| Part of a series on |
| Genetics |
|---|
 |
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, thymine, cytosine, and guanine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery.[1][2]
Knowledge of DNA sequences has become indispensable for basic biological research, DNA Genographic Projects and in numerous applied fields such as medical diagnosis, biotechnology, forensic biology, virology and biological systematics. Comparing healthy and mutated DNA sequences can diagnose different diseases including various cancers,[3] characterize antibody repertoire,[4] and can be used to guide patient treatment.[5] Having a quick way to sequence DNA allows for faster and more individualized medical care to be administered, and for more organisms to be identified and cataloged.[4]
The rapid advancements in DNA sequencing technology have played a crucial role in sequencing complete genomes of humans, as well as numerous animal, plant, and microbial species.

The first DNA sequences were obtained in the early 1970s by academic researchers using laborious methods based on two-dimensional chromatography. Following the development of fluorescence-based sequencing methods with a DNA sequencer,[6] DNA sequencing has become easier and orders of magnitude faster.[7][8]
Applications
[edit]DNA sequencing can be used to determine the sequence of individual genes, larger genetic regions (i.e. clusters of genes or operons), full chromosomes, or entire genomes of any organism. DNA sequencing is also the most efficient way to indirectly sequence RNA or proteins (via their open reading frames). In fact, DNA sequencing has become a key technology in many areas of biology and other sciences such as medicine, forensics, and anthropology. [citation needed]
Molecular biology
[edit]Sequencing is used in molecular biology to study genomes and the proteins they encode. Information obtained using sequencing allows researchers to identify changes in genes and noncoding DNA (including regulatory sequences), associations with diseases and phenotypes, and identify potential drug targets. [citation needed]
Evolutionary biology
[edit]Since DNA is an informative macromolecule in terms of transmission from one generation to another, DNA sequencing is used in evolutionary biology to study how different organisms are related and how they evolved. In February 2021, scientists reported, for the first time, the sequencing of DNA from animal remains, a mammoth in this instance, over a million years old, the oldest DNA sequenced to date.[9][10]
Metagenomics
[edit]The field of metagenomics involves identification of organisms present in a body of water, sewage, dirt, debris filtered from the air, or swab samples from organisms. Knowing which organisms are present in a particular environment is critical to research in ecology, epidemiology, microbiology, and other fields. Sequencing enables researchers to determine which types of microbes may be present in a microbiome, for example. [citation needed]
Virology
[edit]As most viruses are too small to be seen by a light microscope, sequencing is one of the main tools in virology to identify and study the virus.[11] Viral genomes can be based in DNA or RNA. RNA viruses are more time-sensitive for genome sequencing, as they degrade faster in clinical samples.[12] Traditional Sanger sequencing and next-generation sequencing are used to sequence viruses in basic and clinical research, as well as for the diagnosis of emerging viral infections, molecular epidemiology of viral pathogens, and drug-resistance testing. There are more than 2.3 million unique viral sequences in GenBank.[11] In 2019, NGS has surpassed traditional Sanger as the most popular approach for generating viral genomes.[11]
During the 1997 avian influenza outbreak, viral sequencing determined that the influenza sub-type originated through reassortment between quail and poultry. This led to legislation in Hong Kong that prohibited selling live quail and poultry together at market. Viral sequencing can also be used to estimate when a viral outbreak began by using a molecular clock technique.[12]
Medicine
[edit]Medical technicians may sequence genes (or, theoretically, full genomes) from patients to determine if there is risk of genetic diseases. This is a form of genetic testing, though some genetic tests may not involve DNA sequencing. [citation needed]
As of 2013 DNA sequencing was increasingly used to diagnose and treat rare diseases. As more and more genes are identified that cause rare genetic diseases, molecular diagnoses for patients become more mainstream. DNA sequencing allows clinicians to identify genetic diseases, improve disease management, provide reproductive counseling, and more effective therapies.[13] Gene sequencing panels are used to identify multiple potential genetic causes of a suspected disorder.[14]
Also, DNA sequencing may be useful for determining a specific bacteria, to allow for more precise antibiotics treatments, hereby reducing the risk of creating antimicrobial resistance in bacteria populations.[15][16][17][18][19][20]
Forensic investigation
[edit]DNA sequencing may be used along with DNA profiling methods for forensic identification[21] and paternity testing. DNA testing has evolved tremendously in the last few decades to ultimately link a DNA print to what is under investigation. The DNA patterns in fingerprint, saliva, hair follicles, etc. uniquely separate each living organism from another. Testing DNA is a technique which can detect specific genomes in a DNA strand to produce a unique and individualized pattern. [citation needed]
The four canonical bases
[edit]The canonical structure of DNA has four bases: thymine (T), adenine (A), cytosine (C), and guanine (G). DNA sequencing is the determination of the physical order of these bases in a molecule of DNA. However, there are many other bases that may be present in a molecule. In some viruses (specifically, bacteriophage), cytosine may be replaced by hydroxy methyl or hydroxy methyl glucose cytosine.[22] In mammalian DNA, variant bases with methyl groups or phosphosulfate may be found.[23][24] Depending on the sequencing technique, a particular modification, e.g., the 5mC (5-Methylcytosine) common in humans, may or may not be detected.[25]
In almost all organisms, DNA is synthesized in vivo using only the 4 canonical bases; modification that occurs post replication creates other bases like 5 methyl C. However, some bacteriophage can incorporate a non standard base directly.[26]
In addition to modifications, DNA is under constant assault by environmental agents such as UV and Oxygen radicals. At the present time, the presence of such damaged bases is not detected by most DNA sequencing methods, although PacBio has published on this.[27]
History
[edit]Discovery of DNA structure and function
[edit]Deoxyribonucleic acid (DNA) was first discovered and isolated by Friedrich Miescher in 1869, but it remained under-studied for many decades because proteins, rather than DNA, were thought to hold the genetic blueprint to life. This situation changed after 1944 as a result of some experiments by Oswald Avery, Colin MacLeod, and Maclyn McCarty demonstrating that purified DNA could change one strain of bacteria into another. This was the first time that DNA was shown capable of transforming the properties of cells. [citation needed]
In 1953, James Watson and Francis Crick put forward their double-helix model of DNA, based on crystallized X-ray structures being studied by Rosalind Franklin. According to the model, DNA is composed of two strands of nucleotides coiled around each other, linked together by hydrogen bonds and running in opposite directions. Each strand is composed of four complementary nucleotides – adenine (A), cytosine (C), guanine (G) and thymine (T) – with an A on one strand always paired with T on the other, and C always paired with G. They proposed that such a structure allowed each strand to be used to reconstruct the other, an idea central to the passing on of hereditary information between generations.[28]

The foundation for sequencing proteins was first laid by the work of Frederick Sanger who by 1955 had completed the sequence of all the amino acids in insulin, a small protein secreted by the pancreas. This provided the first conclusive evidence that proteins were chemical entities with a specific molecular pattern rather than a random mixture of material suspended in fluid. Sanger's success in sequencing insulin spurred on x-ray crystallographers, including Watson and Crick, who by now were trying to understand how DNA directed the formation of proteins within a cell. Soon after attending a series of lectures given by Frederick Sanger in October 1954, Crick began developing a theory which argued that the arrangement of nucleotides in DNA determined the sequence of amino acids in proteins, which in turn helped determine the function of a protein. He published this theory in 1958.[29]
RNA sequencing
[edit]RNA sequencing was one of the earliest forms of nucleotide sequencing. The major landmark of RNA sequencing is the sequence of the first complete gene and the complete genome of Bacteriophage MS2, identified and published by Walter Fiers and his coworkers at the University of Ghent (Ghent, Belgium), in 1972[30] and 1976.[31] Traditional RNA sequencing methods require the creation of a cDNA molecule which must be sequenced.[32]
Early DNA sequencing methods
[edit]The first method for determining DNA sequences involved a location-specific primer extension strategy established by Ray Wu, a geneticist, at Cornell University in 1970.[33] DNA polymerase catalysis and specific nucleotide labeling, both of which figure prominently in current sequencing schemes, were used to sequence the cohesive ends of lambda phage DNA.[34][35][36] Between 1970 and 1973, Wu, scientist Radha Padmanabhan and colleagues demonstrated that this method can be employed to determine any DNA sequence using synthetic location-specific primers.[37][38][8]
Walter Gilbert, a biochemist, and Allan Maxam, a molecular geneticist, at Harvard also developed sequencing methods, including one for "DNA sequencing by chemical degradation".[39][40] In 1973, Gilbert and Maxam reported the sequence of 24 basepairs using a method known as wandering-spot analysis.[41] Advancements in sequencing were aided by the concurrent development of recombinant DNA technology, allowing DNA samples to be isolated from sources other than viruses.[42]
Two years later in 1975, Frederick Sanger, a biochemist, and Alan Coulson, a genome scientist, developed a method to sequence DNA.[43] The technique known as the "Plus and Minus" method, involved supplying all the components of the DNA but excluding the reaction of one of the four bases needed to complete the DNA.[44]
In 1976, Gilbert and Maxam, invented a method for rapidly sequencing DNA while at Harvard, known as the Maxam–Gilbert sequencing.[45] The technique involved treating radiolabelled DNA with a chemical and using a polyacrylamide gel to determine the sequence.[46]
In 1977, Sanger then adopted a primer-extension strategy to develop more rapid DNA sequencing methods at the MRC Centre, Cambridge, UK. This technique was similar to his "Plus and Minus" strategy, however, it was based upon the selective incorporation of chain-terminating dideoxynucleotides (ddNTPs) by DNA polymerase during in vitro DNA replication.[47][46][48] Sanger published this method in the same year.[49]
Sequencing of full genomes
[edit]
The first full DNA genome to be sequenced was that of bacteriophage φX174 in 1977.[50] Medical Research Council scientists deciphered the complete DNA sequence of the Epstein-Barr virus in 1984, finding it contained 172,282 nucleotides. Completion of the sequence marked a significant turning point in DNA sequencing because it was achieved with no prior genetic profile knowledge of the virus.[51][8]
A non-radioactive method for transferring the DNA molecules of sequencing reaction mixtures onto an immobilizing matrix during electrophoresis was developed by Herbert Pohl and co-workers in the early 1980s.[52][53] Followed by the commercialization of the DNA sequencer "Direct-Blotting-Electrophoresis-System GATC 1500" by GATC Biotech, which was intensively used in the framework of the EU genome-sequencing programme, the complete DNA sequence of the yeast Saccharomyces cerevisiae chromosome II.[54] Leroy E. Hood's laboratory at the California Institute of Technology announced the first semi-automated DNA sequencing machine in 1986.[55] This was followed by Applied Biosystems' marketing of the first fully automated sequencing machine, the ABI 370, in 1987 and by Dupont's Genesis 2000[56] which used a novel fluorescent labeling technique enabling all four dideoxynucleotides to be identified in a single lane. By 1990, the U.S. National Institutes of Health (NIH) had begun large-scale sequencing trials on Mycoplasma capricolum, Escherichia coli, Caenorhabditis elegans, and Saccharomyces cerevisiae at a cost of US$0.75 per base. Meanwhile, sequencing of human cDNA sequences called expressed sequence tags began in Craig Venter's lab, an attempt to capture the coding fraction of the human genome.[57] In 1995, Venter, Hamilton Smith, and colleagues at The Institute for Genomic Research (TIGR) published the first complete genome of a free-living organism, the bacterium Haemophilus influenzae. The circular chromosome contains 1,830,137 bases and its publication in the journal Science[58] marked the first published use of whole-genome shotgun sequencing, eliminating the need for initial mapping efforts.
By 2003, the Human Genome Project's shotgun sequencing methods had been used to produce a draft sequence of the human genome; it had a 92% accuracy.[59][60][61] In 2022, scientists successfully sequenced the last 8% of the human genome. The fully sequenced standard reference gene is called GRCh38.p14, and it contains 3.1 billion base pairs.[62][63]
High-throughput sequencing (HTS) methods
[edit]
Several new methods for DNA sequencing were developed in the mid to late 1990s and were implemented in commercial DNA sequencers by 2000. Together these were called the "next-generation" or "second-generation" sequencing (NGS) methods, in order to distinguish them from the earlier methods, including Sanger sequencing. In contrast to the first generation of sequencing, NGS technology is typically characterized by being highly scalable, allowing the entire genome to be sequenced at once. Usually, this is accomplished by fragmenting the genome into small pieces, randomly sampling for a fragment, and sequencing it using one of a variety of technologies, such as those described below. An entire genome is possible because multiple fragments are sequenced at once (giving it the name "massively parallel" sequencing) in an automated process. [citation needed]
NGS technology has tremendously empowered researchers to look for insights into health, anthropologists to investigate human origins, and is catalyzing the "Personalized Medicine" movement. However, it has also opened the door to more room for error. There are many software tools to carry out the computational analysis of NGS data, often compiled at online platforms such as CSI NGS Portal, each with its own algorithm. Even the parameters within one software package can change the outcome of the analysis. In addition, the large quantities of data produced by DNA sequencing have also required development of new methods and programs for sequence analysis. Several efforts to develop standards in the NGS field have been attempted to address these challenges, most of which have been small-scale efforts arising from individual labs. Most recently, a large, organized, FDA-funded effort has culminated in the BioCompute standard.[65]
On 26 October 1990, Roger Tsien, Pepi Ross, Margaret Fahnestock and Allan J Johnston filed a patent describing stepwise ("base-by-base") sequencing with removable 3' blockers on DNA arrays (blots and single DNA molecules).[66] In 1996, Pål Nyrén and his student Mostafa Ronaghi at the Royal Institute of Technology in Stockholm published their method of pyrosequencing.[67]
On 1 April 1997, Pascal Mayer and Laurent Farinelli submitted patents to the World Intellectual Property Organization describing DNA colony sequencing.[68] The DNA sample preparation and random surface-polymerase chain reaction (PCR) arraying methods described in this patent, coupled to Roger Tsien et al.'s "base-by-base" sequencing method, is now implemented in Illumina's Hi-Seq genome sequencers. [citation needed]
In 1998, Phil Green and Brent Ewing of the University of Washington described their phred quality score for sequencer data analysis,[69] a landmark analysis technique that gained widespread adoption, and which is still the most common metric for assessing the accuracy of a sequencing platform.[70]
Lynx Therapeutics published and marketed massively parallel signature sequencing (MPSS), in 2000. This method incorporated a parallelized, adapter/ligation-mediated, bead-based sequencing technology and served as the first commercially available "next-generation" sequencing method, though no DNA sequencers were sold to independent laboratories.[71]
Basic methods
[edit]Maxam-Gilbert sequencing
[edit]Allan Maxam and Walter Gilbert published a DNA sequencing method in 1977 based on chemical modification of DNA and subsequent cleavage at specific bases.[39] Also known as chemical sequencing, this method allowed purified samples of double-stranded DNA to be used without further cloning. This method's use of radioactive labeling and its technical complexity discouraged extensive use after refinements in the Sanger methods had been made. [citation needed]
Maxam-Gilbert sequencing requires radioactive labeling at one 5' end of the DNA and purification of the DNA fragment to be sequenced. Chemical treatment then generates breaks at a small proportion of one or two of the four nucleotide bases in each of four reactions (G, A+G, C, C+T). The concentration of the modifying chemicals is controlled to introduce on average one modification per DNA molecule. Thus a series of labeled fragments is generated, from the radiolabeled end to the first "cut" site in each molecule. The fragments in the four reactions are electrophoresed side by side in denaturing acrylamide gels for size separation. To visualize the fragments, the gel is exposed to X-ray film for autoradiography, yielding a series of dark bands each corresponding to a radiolabeled DNA fragment, from which the sequence may be inferred.[39]
This method is mostly obsolete as of 2023.[72]
Chain-termination methods
[edit]The chain-termination method developed by Frederick Sanger and coworkers in 1977 soon became the method of choice, owing to its relative ease and reliability.[49][73] When invented, the chain-terminator method used fewer toxic chemicals and lower amounts of radioactivity than the Maxam and Gilbert method. Because of its comparative ease, the Sanger method was soon automated and was the method used in the first generation of DNA sequencers. [citation needed]
Sanger sequencing is the method which prevailed from the 1980s until the mid-2000s. Over that period, great advances were made in the technique, such as fluorescent labelling, capillary electrophoresis, and general automation. These developments allowed much more efficient sequencing, leading to lower costs. The Sanger method, in mass production form, is the technology which produced the first human genome in 2001, ushering in the age of genomics. However, later in the decade, radically different approaches reached the market, bringing the cost per genome down from $100 million in 2001 to $10,000 in 2011.[74]
Sequencing by synthesis
[edit]The objective for sequential sequencing by synthesis (SBS) is to determine the sequencing of a DNA sample by detecting the incorporation of a nucleotide by a DNA polymerase. An engineered polymerase is used to synthesize a copy of a single strand of DNA and the incorporation of each nucleotide is monitored. The principle of real-time sequencing by synthesis was first described in 1993[75] with improvements published some years later.[76] The key parts are highly similar for all embodiments of SBS and includes (1) amplification of DNA (to enhance the subsequent signal) and attach the DNA to be sequenced to a solid support, (2) generation of single stranded DNA on the solid support, (3) incorporation of nucleotides using an engineered polymerase and (4) real-time detection of the incorporation of nucleotide The steps 3-4 are repeated and the sequence is assembled from the signals obtained in step 4. This principle of real-time sequencing-by-synthesis has been used for almost all massive parallel sequencing instruments, including 454, PacBio, IonTorrent, Illumina and MGI. [citation needed]
Large-scale sequencing and de novo sequencing
[edit]
Large-scale sequencing often aims at sequencing very long DNA pieces, such as whole chromosomes, although large-scale sequencing can also be used to generate very large numbers of short sequences, such as found in phage display. For longer targets such as chromosomes, common approaches consist of cutting (with restriction enzymes) or shearing (with mechanical forces) large DNA fragments into shorter DNA fragments. The fragmented DNA may then be cloned into a DNA vector and amplified in a bacterial host such as Escherichia coli. Short DNA fragments purified from individual bacterial colonies are individually sequenced and assembled electronically into one long, contiguous sequence. Studies have shown that adding a size selection step to collect DNA fragments of uniform size can improve sequencing efficiency and accuracy of the genome assembly. In these studies, automated sizing has proven to be more reproducible and precise than manual gel sizing.[77][78][79]
The term "de novo sequencing" specifically refers to methods used to determine the sequence of DNA with no previously known sequence. De novo translates from Latin as "from the beginning". Gaps in the assembled sequence may be filled by primer walking. The different strategies have different tradeoffs in speed and accuracy; shotgun methods are often used for sequencing large genomes, but its assembly is complex and difficult, particularly with sequence repeats often causing gaps in genome assembly. [citation needed]
Most sequencing approaches use an in vitro cloning step to amplify individual DNA molecules, because their molecular detection methods are not sensitive enough for single molecule sequencing. Emulsion PCR[80] isolates individual DNA molecules along with primer-coated beads in aqueous droplets within an oil phase. A polymerase chain reaction (PCR) then coats each bead with clonal copies of the DNA molecule followed by immobilization for later sequencing. Emulsion PCR is used in the methods developed by Marguilis et al. (commercialized by 454 Life Sciences), Shendure and Porreca et al. (also known as "polony sequencing") and SOLiD sequencing, (developed by Agencourt, later Applied Biosystems, now Life Technologies).[81][82][83] Emulsion PCR is also used in the GemCode and Chromium platforms developed by 10x Genomics.[84]
Shotgun sequencing
[edit]Shotgun sequencing is a sequencing method designed for analysis of DNA sequences longer than 1000 base pairs, up to and including entire chromosomes. This method requires the target DNA to be broken into random fragments. After sequencing individual fragments using the chain termination method, the sequences can be reassembled on the basis of their overlapping regions.[85]
High-throughput methods
[edit]
High-throughput sequencing, which includes next-generation "short-read" and third-generation "long-read" sequencing methods,[nt 1] applies to exome sequencing, genome sequencing, genome resequencing, transcriptome profiling (RNA-Seq), DNA-protein interactions (ChIP-sequencing), and epigenome characterization.[86]
The high demand for low-cost sequencing has driven the development of high-throughput sequencing technologies that parallelize the sequencing process, producing thousands or millions of sequences concurrently.[87][88][89] High-throughput sequencing technologies are intended to lower the cost of DNA sequencing beyond what is possible with standard dye-terminator methods.[90] In ultra-high-throughput sequencing as many as 500,000 sequencing-by-synthesis operations may be run in parallel.[91][92][93] Such technologies led to the ability to sequence an entire human genome in as little as one day.[94] As of 2019[update], corporate leaders in the development of high-throughput sequencing products included Illumina, Qiagen and ThermoFisher Scientific.[94]
| Method | Read length | Accuracy (single read not consensus) | Reads per run | Time per run | Cost per 1 billion bases (in US$) | Advantages | Disadvantages |
|---|---|---|---|---|---|---|---|
| Single-molecule real-time sequencing (Pacific Biosciences) | 30,000 bp (N50); | 87% raw-read accuracy[100] | 4,000,000 per Sequel 2 SMRT cell, 100–200 gigabases[97][101][102] | 30 minutes to 20 hours[97][103] | $7.2-$43.3 | Fast. Detects 4mC, 5mC, 6mA.[104] | Moderate throughput. Equipment can be very expensive. |
| Ion semiconductor (Ion Torrent sequencing) | up to 600 bp[105] | 99.6%[106] | up to 80 million | 2 hours | $66.8-$950 | Less expensive equipment. Fast. | Homopolymer errors. |
| Pyrosequencing (454) | 700 bp | 99.9% | 1 million | 24 hours | $10,000 | Long read size. Fast. | Runs are expensive. Homopolymer errors. |
| Sequencing by synthesis (Illumina) | MiniSeq, NextSeq: 75–300 bp;
MiSeq: 50–600 bp; HiSeq 2500: 50–500 bp; HiSeq 3/4000: 50–300 bp; HiSeq X: 300 bp |
99.9% (Phred30) | MiniSeq/MiSeq: 1–25 Million;
NextSeq: 130-00 Million; HiSeq 2500: 300 million – 2 billion; HiSeq 3/4000 2.5 billion; HiSeq X: 3 billion |
1 to 11 days, depending upon sequencer and specified read length[107] | $5 to $150 | Potential for high sequence yield, depending upon sequencer model and desired application. | Equipment can be very expensive. Requires high concentrations of DNA. |
| Combinatorial probe anchor synthesis (cPAS- BGI/MGI) | BGISEQ-50: 35-50bp;
MGISEQ 200: 50-200bp; BGISEQ-500, MGISEQ-2000: 50-300bp[108] |
99.9% (Phred30) | BGISEQ-50: 160M;
MGISEQ 200: 300M; BGISEQ-500: 1300M per flow cell; MGISEQ-2000: 375M FCS flow cell, 1500M FCL flow cell per flow cell. |
1 to 9 days depending on instrument, read length and number of flow cells run at a time. | $5– $120 | ||
| Sequencing by ligation (SOLiD sequencing) | 50+35 or 50+50 bp | 99.9% | 1.2 to 1.4 billion | 1 to 2 weeks | $60–130 | Low cost per base. | Slower than other methods. Has issues sequencing palindromic sequences.[109] |
| Nanopore Sequencing | Dependent on library preparation, not the device, so user chooses read length (up to 2,272,580 bp reported[110]). | ~92–97% single read | dependent on read length selected by user | data streamed in real time. Choose 1 min to 48 hrs | $7–100 | Longest individual reads. Accessible user community. Portable (Palm sized). | Lower throughput than other machines, Single read accuracy in 90s. |
| GenapSys Sequencing | Around 150 bp single-end | 99.9% (Phred30) | 1 to 16 million | Around 24 hours | $667 | Low-cost of instrument ($10,000) | |
| Chain termination (Sanger sequencing) | 400 to 900 bp | 99.9% | N/A | 20 minutes to 3 hours | $2,400,000 | Useful for many applications. | More expensive and impractical for larger sequencing projects. This method also requires the time-consuming step of plasmid cloning or PCR. |
Long-read sequencing methods
[edit]Single molecule real time (SMRT) sequencing
[edit]SMRT sequencing is based on the sequencing by synthesis approach. The DNA is synthesized in zero-mode wave-guides (ZMWs) – small well-like containers with the capturing tools located at the bottom of the well. The sequencing is performed with use of unmodified polymerase (attached to the ZMW bottom) and fluorescently labelled nucleotides flowing freely in the solution. The wells are constructed in a way that only the fluorescence occurring by the bottom of the well is detected. The fluorescent label is detached from the nucleotide upon its incorporation into the DNA strand, leaving an unmodified DNA strand. According to Pacific Biosciences (PacBio), the SMRT technology developer, this methodology allows detection of nucleotide modifications (such as cytosine methylation). This happens through the observation of polymerase kinetics. This approach allows reads of 20,000 nucleotides or more, with average read lengths of 5 kilobases.[101][111] In 2015, Pacific Biosciences announced the launch of a new sequencing instrument called the Sequel System, with 1 million ZMWs compared to 150,000 ZMWs in the PacBio RS II instrument.[112][113] SMRT sequencing is referred to as "third-generation" or "long-read" sequencing. [citation needed]
Nanopore DNA sequencing
[edit]The DNA passing through the nanopore changes its ion current. This change is dependent on the shape, size and length of the DNA sequence. Each type of the nucleotide blocks the ion flow through the pore for a different period of time. The method does not require modified nucleotides and is performed in real time. Nanopore sequencing is referred to as "third-generation" or "long-read" sequencing, along with SMRT sequencing. [citation needed]
Early industrial research into this method was based on a technique called 'exonuclease sequencing', where the readout of electrical signals occurred as nucleotides passed by alpha(α)-hemolysin pores covalently bound with cyclodextrin.[114] However the subsequent commercial method, 'strand sequencing', sequenced DNA bases in an intact strand. [citation needed]
Two main areas of nanopore sequencing in development are solid state nanopore sequencing, and protein based nanopore sequencing. Protein nanopore sequencing utilizes membrane protein complexes such as α-hemolysin, MspA (Mycobacterium smegmatis Porin A) or CssG, which show great promise given their ability to distinguish between individual and groups of nucleotides.[115] In contrast, solid-state nanopore sequencing utilizes synthetic materials such as silicon nitride and aluminum oxide and it is preferred for its superior mechanical ability and thermal and chemical stability.[116] The fabrication method is essential for this type of sequencing given that the nanopore array can contain hundreds of pores with diameters smaller than eight nanometers.[115]
The concept originated from the idea that single stranded DNA or RNA molecules can be electrophoretically driven in a strict linear sequence through a biological pore that can be less than eight nanometers, and can be detected given that the molecules release an ionic current while moving through the pore. The pore contains a detection region capable of recognizing different bases, with each base generating various time specific signals corresponding to the sequence of bases as they cross the pore which are then evaluated.[116] Precise control over the DNA transport through the pore is crucial for success. Various enzymes such as exonucleases and polymerases have been used to moderate this process by positioning them near the pore's entrance.[117]
Short-read sequencing methods
[edit]Massively parallel signature sequencing (MPSS)
[edit]The first of the high-throughput sequencing technologies, massively parallel signature sequencing (or MPSS, also called next generation sequencing), was developed in the 1990s at Lynx Therapeutics, a company founded in 1992 by Sydney Brenner and Sam Eletr. MPSS was a bead-based method that used a complex approach of adapter ligation followed by adapter decoding, reading the sequence in increments of four nucleotides. This method made it susceptible to sequence-specific bias or loss of specific sequences. Because the technology was so complex, MPSS was only performed 'in-house' by Lynx Therapeutics and no DNA sequencing machines were sold to independent laboratories. Lynx Therapeutics merged with Solexa (later acquired by Illumina) in 2004, leading to the development of sequencing-by-synthesis, a simpler approach acquired from Manteia Predictive Medicine, which rendered MPSS obsolete. However, the essential properties of the MPSS output were typical of later high-throughput data types, including hundreds of thousands of short DNA sequences. In the case of MPSS, these were typically used for sequencing cDNA for measurements of gene expression levels.[71]
Polony sequencing
[edit]The polony sequencing method, developed in the laboratory of George M. Church at Harvard, was among the first high-throughput sequencing systems and was used to sequence a full E. coli genome in 2005.[82] It combined an in vitro paired-tag library with emulsion PCR, an automated microscope, and ligation-based sequencing chemistry to sequence an E. coli genome at an accuracy of >99.9999% and a cost approximately 1/9 that of Sanger sequencing.[82] The technology was licensed to Agencourt Biosciences, subsequently spun out into Agencourt Personal Genomics, and eventually incorporated into the Applied Biosystems SOLiD platform. Applied Biosystems was later acquired by Life Technologies, now part of Thermo Fisher Scientific. [citation needed]
454 pyrosequencing
[edit]A parallelized version of pyrosequencing was developed by 454 Life Sciences, which has since been acquired by Roche Diagnostics. The method amplifies DNA inside water droplets in an oil solution (emulsion PCR), with each droplet containing a single DNA template attached to a single primer-coated bead that then forms a clonal colony. The sequencing machine contains many picoliter-volume wells each containing a single bead and sequencing enzymes. Pyrosequencing uses luciferase to generate light for detection of the individual nucleotides added to the nascent DNA, and the combined data are used to generate sequence reads.[81] This technology provides intermediate read length and price per base compared to Sanger sequencing on one end and Solexa and SOLiD on the other.[90]
Illumina (Solexa) sequencing
[edit]Solexa, now part of Illumina, was founded by Shankar Balasubramanian and David Klenerman in 1998, and developed a sequencing method based on reversible dye-terminators technology, and engineered polymerases.[118] The reversible terminated chemistry concept was invented by Bruno Canard and Simon Sarfati at the Pasteur Institute in Paris.[119][120] It was developed internally at Solexa by those named on the relevant patents. In 2004, Solexa acquired the company Manteia Predictive Medicine in order to gain a massively parallel sequencing technology invented in 1997 by Pascal Mayer and Laurent Farinelli.[68] It is based on "DNA clusters" or "DNA colonies", which involves the clonal amplification of DNA on a surface. The cluster technology was co-acquired with Lynx Therapeutics of California. Solexa Ltd. later merged with Lynx to form Solexa Inc. [citation needed]


In this method, DNA molecules and primers are first attached on a slide or flow cell and amplified with polymerase so that local clonal DNA colonies, later coined "DNA clusters", are formed. To determine the sequence, four types of reversible terminator bases (RT-bases) are added and non-incorporated nucleotides are washed away. A camera takes images of the fluorescently labeled nucleotides. Then the dye, along with the terminal 3' blocker, is chemically removed from the DNA, allowing for the next cycle to begin. Unlike pyrosequencing, the DNA chains are extended one nucleotide at a time and image acquisition can be performed at a delayed moment, allowing for very large arrays of DNA colonies to be captured by sequential images taken from a single camera. [citation needed]

Decoupling the enzymatic reaction and the image capture allows for optimal throughput and theoretically unlimited sequencing capacity. With an optimal configuration, the ultimately reachable instrument throughput is thus dictated solely by the analog-to-digital conversion rate of the camera, multiplied by the number of cameras and divided by the number of pixels per DNA colony required for visualizing them optimally (approximately 10 pixels/colony). In 2012, with cameras operating at more than 10 MHz A/D conversion rates and available optics, fluidics and enzymatics, throughput can be multiples of 1 million nucleotides/second, corresponding roughly to 1 human genome equivalent at 1x coverage per hour per instrument, and 1 human genome re-sequenced (at approx. 30x) per day per instrument (equipped with a single camera).[121]
Combinatorial probe anchor synthesis (cPAS)
[edit]This method is an upgraded modification to combinatorial probe anchor ligation technology (cPAL) described by Complete Genomics[122] which has since become part of Chinese genomics company BGI in 2013.[123] The two companies have refined the technology to allow for longer read lengths, reaction time reductions and faster time to results. In addition, data are now generated as contiguous full-length reads in the standard FASTQ file format and can be used as-is in most short-read-based bioinformatics analysis pipelines.[124][125]
The two technologies that form the basis for this high-throughput sequencing technology are DNA nanoballs (DNB) and patterned arrays for nanoball attachment to a solid surface.[122] DNA nanoballs are simply formed by denaturing double stranded, adapter ligated libraries and ligating the forward strand only to a splint oligonucleotide to form a ssDNA circle. Faithful copies of the circles containing the DNA insert are produced utilizing Rolling Circle Amplification that generates approximately 300–500 copies. The long strand of ssDNA folds upon itself to produce a three-dimensional nanoball structure that is approximately 220 nm in diameter. Making DNBs replaces the need to generate PCR copies of the library on the flow cell and as such can remove large proportions of duplicate reads, adapter-adapter ligations and PCR induced errors.[124][126]

The patterned array of positively charged spots is fabricated through photolithography and etching techniques followed by chemical modification to generate a sequencing flow cell. Each spot on the flow cell is approximately 250 nm in diameter, are separated by 700 nm (centre to centre) and allows easy attachment of a single negatively charged DNB to the flow cell and thus reducing under or over-clustering on the flow cell.[122][127]
Sequencing is then performed by addition of an oligonucleotide probe that attaches in combination to specific sites within the DNB. The probe acts as an anchor that then allows one of four single reversibly inactivated, labelled nucleotides to bind after flowing across the flow cell. Unbound nucleotides are washed away before laser excitation of the attached labels then emit fluorescence and signal is captured by cameras that is converted to a digital output for base calling. The attached base has its terminator and label chemically cleaved at completion of the cycle. The cycle is repeated with another flow of free, labelled nucleotides across the flow cell to allow the next nucleotide to bind and have its signal captured. This process is completed a number of times (usually 50 to 300 times) to determine the sequence of the inserted piece of DNA at a rate of approximately 40 million nucleotides per second as of 2018.[citation needed]
SOLiD sequencing
[edit]
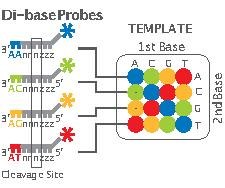
Applied Biosystems' (now a Life Technologies brand) SOLiD technology employs sequencing by ligation. Here, a pool of all possible oligonucleotides of a fixed length are labeled according to the sequenced position. Oligonucleotides are annealed and ligated; the preferential ligation by DNA ligase for matching sequences results in a signal informative of the nucleotide at that position. Each base in the template is sequenced twice, and the resulting data are decoded according to the 2 base encoding scheme used in this method. Before sequencing, the DNA is amplified by emulsion PCR. The resulting beads, each containing single copies of the same DNA molecule, are deposited on a glass slide.[128] The result is sequences of quantities and lengths comparable to Illumina sequencing.[90] This sequencing by ligation method has been reported to have some issue sequencing palindromic sequences.[109]
Ion Torrent semiconductor sequencing
[edit]Ion Torrent Systems Inc. (now owned by Life Technologies) developed a system based on using standard sequencing chemistry, but with a novel, semiconductor-based detection system. This method of sequencing is based on the detection of hydrogen ions that are released during the polymerisation of DNA, as opposed to the optical methods used in other sequencing systems. A microwell containing a template DNA strand to be sequenced is flooded with a single type of nucleotide. If the introduced nucleotide is complementary to the leading template nucleotide it is incorporated into the growing complementary strand. This causes the release of a hydrogen ion that triggers a hypersensitive ion sensor, which indicates that a reaction has occurred. If homopolymer repeats are present in the template sequence, multiple nucleotides will be incorporated in a single cycle. This leads to a corresponding number of released hydrogens and a proportionally higher electronic signal.[129]

DNA nanoball sequencing
[edit]DNA nanoball sequencing is a type of high throughput sequencing technology used to determine the entire genomic sequence of an organism. The company Complete Genomics uses this technology to sequence samples submitted by independent researchers. The method uses rolling circle replication to amplify small fragments of genomic DNA into DNA nanoballs. Unchained sequencing by ligation is then used to determine the nucleotide sequence.[130] This method of DNA sequencing allows large numbers of DNA nanoballs to be sequenced per run and at low reagent costs compared to other high-throughput sequencing platforms.[131] However, only short sequences of DNA are determined from each DNA nanoball which makes mapping the short reads to a reference genome difficult.[130]
Heliscope single molecule sequencing
[edit]Heliscope sequencing is a method of single-molecule sequencing developed by Helicos Biosciences. It uses DNA fragments with added poly-A tail adapters which are attached to the flow cell surface. The next steps involve extension-based sequencing with cyclic washes of the flow cell with fluorescently labeled nucleotides (one nucleotide type at a time, as with the Sanger method). The reads are performed by the Heliscope sequencer.[132][133] The reads are short, averaging 35 bp.[134] What made this technology especially novel was that it was the first of its class to sequence non-amplified DNA, thus preventing any read errors associated with amplification steps.[46] In 2009 a human genome was sequenced using the Heliscope, however in 2012 the company went bankrupt.[135]
Microfluidic systems
[edit]There are two main microfluidic systems that are used to sequence DNA; droplet based microfluidics and digital microfluidics. Microfluidic devices solve many of the current limitations of current sequencing arrays. [citation needed]
Abate et al. studied the use of droplet-based microfluidic devices for DNA sequencing.[4] These devices have the ability to form and process picoliter sized droplets at the rate of thousands per second. The devices were created from polydimethylsiloxane (PDMS) and used Forster resonance energy transfer, FRET assays to read the sequences of DNA encompassed in the droplets. Each position on the array tested for a specific 15 base sequence.[4]
Fair et al. used digital microfluidic devices to study DNA pyrosequencing.[136] Significant advantages include the portability of the device, reagent volume, speed of analysis, mass manufacturing abilities, and high throughput. This study provided a proof of concept showing that digital devices can be used for pyrosequencing; the study included using synthesis, which involves the extension of the enzymes and addition of labeled nucleotides.[136]
Boles et al. also studied pyrosequencing on digital microfluidic devices.[137] They used an electro-wetting device to create, mix, and split droplets. The sequencing uses a three-enzyme protocol and DNA templates anchored with magnetic beads. The device was tested using two protocols and resulted in 100% accuracy based on raw pyrogram levels. The advantages of these digital microfluidic devices include size, cost, and achievable levels of functional integration.[137]
DNA sequencing research, using microfluidics, also has the ability to be applied to the sequencing of RNA, using similar droplet microfluidic techniques, such as the method, inDrops.[138] This shows that many of these DNA sequencing techniques will be able to be applied further and be used to understand more about genomes and transcriptomes. [citation needed]
Methods in development
[edit]DNA sequencing methods currently under development include reading the sequence as a DNA strand transits through nanopores (a method that is now commercial but subsequent generations such as solid-state nanopores are still in development),[139][140] and microscopy-based techniques, such as atomic force microscopy or transmission electron microscopy that are used to identify the positions of individual nucleotides within long DNA fragments (>5,000 bp) by nucleotide labeling with heavier elements (e.g., halogens) for visual detection and recording.[141][142] Third generation technologies aim to increase throughput and decrease the time to result and cost by eliminating the need for excessive reagents and harnessing the processivity of DNA polymerase.[143]
Tunnelling currents DNA sequencing
[edit]Another approach uses measurements of the electrical tunnelling currents across single-strand DNA as it moves through a channel. Depending on its electronic structure, each base affects the tunnelling current differently,[144] allowing differentiation between different bases.[145]
The use of tunnelling currents has the potential to sequence orders of magnitude faster than ionic current methods and the sequencing of several DNA oligomers and micro-RNA has already been achieved.[146]
Sequencing by hybridization
[edit]Sequencing by hybridization is a non-enzymatic method that uses a DNA microarray. A single pool of DNA whose sequence is to be determined is fluorescently labeled and hybridized to an array containing known sequences. Strong hybridization signals from a given spot on the array identifies its sequence in the DNA being sequenced.[147]
This method of sequencing utilizes binding characteristics of a library of short single stranded DNA molecules (oligonucleotides), also called DNA probes, to reconstruct a target DNA sequence. Non-specific hybrids are removed by washing and the target DNA is eluted.[148] Hybrids are re-arranged such that the DNA sequence can be reconstructed. The benefit of this sequencing type is its ability to capture a large number of targets with a homogenous coverage.[149] A large number of chemicals and starting DNA is usually required. However, with the advent of solution-based hybridization, much less equipment and chemicals are necessary.[148]
Sequencing with mass spectrometry
[edit]Mass spectrometry may be used to determine DNA sequences. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, or MALDI-TOF MS, has specifically been investigated as an alternative method to gel electrophoresis for visualizing DNA fragments. With this method, DNA fragments generated by chain-termination sequencing reactions are compared by mass rather than by size. The mass of each nucleotide is different from the others and this difference is detectable by mass spectrometry. Single-nucleotide mutations in a fragment can be more easily detected with MS than by gel electrophoresis alone. MALDI-TOF MS can more easily detect differences between RNA fragments, so researchers may indirectly sequence DNA with MS-based methods by converting it to RNA first.[150]
The higher resolution of DNA fragments permitted by MS-based methods is of special interest to researchers in forensic science, as they may wish to find single-nucleotide polymorphisms in human DNA samples to identify individuals. These samples may be highly degraded so forensic researchers often prefer mitochondrial DNA for its higher stability and applications for lineage studies. MS-based sequencing methods have been used to compare the sequences of human mitochondrial DNA from samples in a Federal Bureau of Investigation database[151] and from bones found in mass graves of World War I soldiers.[152]
Early chain-termination and TOF MS methods demonstrated read lengths of up to 100 base pairs.[153] Researchers have been unable to exceed this average read size; like chain-termination sequencing alone, MS-based DNA sequencing may not be suitable for large de novo sequencing projects. Even so, a 2010 study did use the short sequence reads and mass spectroscopy to compare single-nucleotide polymorphisms in pathogenic Streptococcus strains.[154]
Microfluidic Sanger sequencing
[edit]In microfluidic Sanger sequencing the entire thermocycling amplification of DNA fragments as well as their separation by electrophoresis is done on a single glass wafer (approximately 10 cm in diameter) thus reducing the reagent usage as well as cost.[155] In some instances researchers have shown that they can increase the throughput of conventional sequencing through the use of microchips.[156] Research will still need to be done in order to make this use of technology effective. [citation needed]
Microscopy-based techniques
[edit]This approach directly visualizes the sequence of DNA molecules using electron microscopy. The first identification of DNA base pairs within intact DNA molecules by enzymatically incorporating modified bases, which contain atoms of increased atomic number, direct visualization and identification of individually labeled bases within a synthetic 3,272 base-pair DNA molecule and a 7,249 base-pair viral genome has been demonstrated.[157]
RNAP sequencing
[edit]This method is based on use of RNA polymerase (RNAP), which is attached to a polystyrene bead. One end of DNA to be sequenced is attached to another bead, with both beads being placed in optical traps. RNAP motion during transcription brings the beads in closer and their relative distance changes, which can then be recorded at a single nucleotide resolution. The sequence is deduced based on the four readouts with lowered concentrations of each of the four nucleotide types, similarly to the Sanger method.[158] A comparison is made between regions and sequence information is deduced by comparing the known sequence regions to the unknown sequence regions.[158]
In vitro virus high-throughput sequencing
[edit]A method has been developed to analyze full sets of protein interactions using a combination of 454 pyrosequencing and an in vitro virus mRNA display method. Specifically, this method covalently links proteins of interest to the mRNAs encoding them, then detects the mRNA pieces using reverse transcription PCRs. The mRNA may then be amplified and sequenced. The combined method was titled IVV-HiTSeq and can be performed under cell-free conditions, though its results may not be representative of in vivo conditions.[159]
Market share
[edit]While there are many different ways to sequence DNA, only a few dominate the market. In 2022, Illumina had about 80% of the market; the rest of the market is taken by only a few players (PacBio, Oxford, 454, MGI)[160]
Sample preparation
[edit]The success of any DNA sequencing protocol relies upon the DNA or RNA sample extraction and preparation from the biological material of interest. [citation needed]
- A successful DNA extraction will yield a DNA sample with long, non-degraded strands.
- A successful RNA extraction will yield a RNA sample that should be converted to complementary DNA (cDNA) using reverse transcriptase—a DNA polymerase that synthesizes a complementary DNA based on existing strands of RNA in a PCR-like manner.[161] Complementary DNA can then be processed the same way as genomic DNA.
After DNA or RNA extraction, samples may require further preparation depending on the sequencing method. For Sanger sequencing, either cloning procedures or PCR are required prior to sequencing. In the case of next-generation sequencing methods, library preparation is required before processing.[162] Assessing the quality and quantity of nucleic acids both after extraction and after library preparation identifies degraded, fragmented, and low-purity samples and yields high-quality sequencing data.[163]
Development initiatives
[edit]
In October 2006, the X Prize Foundation established an initiative to promote the development of full genome sequencing technologies, called the Archon X Prize, intending to award $10 million to "the first Team that can build a device and use it to sequence 100 human genomes within 10 days or less, with an accuracy of no more than one error in every 100,000 bases sequenced, with sequences accurately covering at least 98% of the genome, and at a recurring cost of no more than $10,000 (US) per genome."[164]
Each year the National Human Genome Research Institute, or NHGRI, promotes grants for new research and developments in genomics. 2010 grants and 2011 candidates include continuing work in microfluidic, polony and base-heavy sequencing methodologies.[165]
Computational challenges
[edit]The sequencing technologies described here produce raw data that needs to be assembled into longer sequences such as complete genomes (sequence assembly). There are many computational challenges to achieve this, such as the evaluation of the raw sequence data which is done by programs and algorithms such as Phred and Phrap. Other challenges have to deal with repetitive sequences that often prevent complete genome assemblies because they occur in many places of the genome. As a consequence, many sequences may not be assigned to particular chromosomes. The production of raw sequence data is only the beginning of its detailed bioinformatical analysis.[166] Yet new methods for sequencing and correcting sequencing errors were developed.[167]
Read trimming
[edit]Sometimes, the raw reads produced by the sequencer are correct and precise only in a fraction of their length. Using the entire read may introduce artifacts in the downstream analyses like genome assembly, SNP calling, or gene expression estimation. Two classes of trimming programs have been introduced, based on the window-based or the running-sum classes of algorithms.[168] This is a partial list of the trimming algorithms currently available, specifying the algorithm class they belong to:
| Name of algorithm | Type of algorithm |
|---|---|
| Cutadapt[169] | Running sum |
| ConDeTri[170] | Window based |
| ERNE-FILTER[171] | Running sum |
| FASTX quality trimmer | Window based |
| PRINSEQ[172] | Window based |
| Trimmomatic[173] | Window based |
| SolexaQA[174] | Window based |
| SolexaQA-BWA | Running sum |
| Sickle | Window based |
Ethical issues
[edit]Human genetics have been included within the field of bioethics since the early 1970s[175] and the growth in the use of DNA sequencing (particularly high-throughput sequencing) has introduced a number of ethical issues. One key issue is the ownership of an individual's DNA and the data produced when that DNA is sequenced.[176] Regarding the DNA molecule itself, the leading legal case on this topic, Moore v. Regents of the University of California (1990) ruled that individuals have no property rights to discarded cells or any profits made using these cells (for instance, as a patented cell line). However, individuals have a right to informed consent regarding removal and use of cells. Regarding the data produced through DNA sequencing, Moore gives the individual no rights to the information derived from their DNA.[176]
As DNA sequencing becomes more widespread, the storage, security and sharing of genomic data has also become more important.[176][177] For instance, one concern is that insurers may use an individual's genomic data to modify their quote, depending on the perceived future health of the individual based on their DNA.[177][178] In May 2008, the Genetic Information Nondiscrimination Act (GINA) was signed in the United States, prohibiting discrimination on the basis of genetic information with respect to health insurance and employment.[179][180] In 2012, the US Presidential Commission for the Study of Bioethical Issues reported that existing privacy legislation for DNA sequencing data such as GINA and the Health Insurance Portability and Accountability Act were insufficient, noting that whole-genome sequencing data was particularly sensitive, as it could be used to identify not only the individual from which the data was created, but also their relatives.[181][182]
In most of the United States, DNA that is "abandoned", such as that found on a licked stamp or envelope, coffee cup, cigarette, chewing gum, household trash, or hair that has fallen on a public sidewalk, may legally be collected and sequenced by anyone, including the police, private investigators, political opponents, or people involved in paternity disputes. As of 2013, eleven states have laws that can be interpreted to prohibit "DNA theft".[183]
Ethical issues have also been raised by the increasing use of genetic variation screening, both in newborns, and in adults by companies such as 23andMe.[184][185] It has been asserted that screening for genetic variations can be harmful, increasing anxiety in individuals who have been found to have an increased risk of disease.[186] For example, in one case noted in Time, doctors screening an ill baby for genetic variants chose not to inform the parents of an unrelated variant linked to dementia due to the harm it would cause to the parents.[187] However, a 2011 study in The New England Journal of Medicine has shown that individuals undergoing disease risk profiling did not show increased levels of anxiety.[186] Also, the development of Next Generation sequencing technologies such as Nanopore based sequencing has also raised further ethical concerns.[188]
See also
[edit]- Bioinformatics – Computational analysis of large, complex sets of biological data
- Cancer genome sequencing
- Circular consensus sequencing
- DNA computing – Computing using molecular biology hardware
- DNA field-effect transistor
- DNA sequencing theory – Biological theory
- DNA sequencer – Scientific instrument that automates the DNA sequencing process
- Genographic Project – Citizen science project
- Genome project – Scientific endeavours to determine the complete genome sequence of an organism
- Genome sequencing of endangered species – DNA testing for endangerment assessment
- Genome skimming – Method of genome sequencing
- IsoBase – Database for identifying functionally related proteins
- Linked-read sequencing
- Jumping library
- Nucleic acid sequence – Succession of nucleotides in a nucleic acid
- Multiplex ligation-dependent probe amplification
- Personalized medicine – Medical model that tailors medical practices to the individual patient
- Protein sequencing – Sequencing of amino acid arrangement in a protein
- Sequence mining – Data mining technique
- Sequence profiling tool
- Sequencing by hybridization
- Sequencing by ligation
- TIARA (database) – Database of personal genomics information
- Transmission electron microscopy DNA sequencing – Single-molecule sequencing technology
Notes
[edit]- ^ "Next-generation" remains in broad use as of 2019. For instance, Straiton J, Free T, Sawyer A, Martin J (February 2019). "From Sanger Sequencing to Genome Databases and Beyond". BioTechniques. 66 (2): 60–63. doi:10.2144/btn-2019-0011. PMID 30744413.
Next-generation sequencing (NGS) technologies have revolutionized genomic research. (opening sentence of the article)
References
[edit]- ^ "Introducing 'dark DNA' – the phenomenon that could change how we think about evolution". 24 August 2017.
- ^ Behjati S, Tarpey PS (December 2013). "What is next generation sequencing?". Archives of Disease in Childhood: Education and Practice Edition. 98 (6): 236–8. doi:10.1136/archdischild-2013-304340. PMC 3841808. PMID 23986538.
- ^ Chmielecki J, Meyerson M (14 January 2014). "DNA sequencing of cancer: what have we learned?". Annual Review of Medicine. 65 (1): 63–79. doi:10.1146/annurev-med-060712-200152. PMID 24274178.
- ^ a b c d Abate AR, Hung T, Sperling RA, Mary P, Rotem A, Agresti JJ, et al. (December 2013). "DNA sequence analysis with droplet-based microfluidics". Lab on a Chip. 13 (24): 4864–9. doi:10.1039/c3lc50905b. PMC 4090915. PMID 24185402.
- ^ Pekin D, Skhiri Y, Baret JC, Le Corre D, Mazutis L, Salem CB, et al. (July 2011). "Quantitative and sensitive detection of rare mutations using droplet-based microfluidics". Lab on a Chip. 11 (13): 2156–66. doi:10.1039/c1lc20128j. PMID 21594292.
- ^ Olsvik O, Wahlberg J, Petterson B, Uhlén M, Popovic T, Wachsmuth IK, Fields PI (January 1993). "Use of automated sequencing of polymerase chain reaction-generated amplicons to identify three types of cholera toxin subunit B in Vibrio cholerae O1 strains". J. Clin. Microbiol. 31 (1): 22–25. doi:10.1128/JCM.31.1.22-25.1993. PMC 262614. PMID 7678018.

- ^ Pettersson E, Lundeberg J, Ahmadian A (February 2009). "Generations of sequencing technologies". Genomics. 93 (2): 105–11. doi:10.1016/j.ygeno.2008.10.003. PMID 18992322.
- ^ a b c Jay E, Bambara R, Padmanabhan R, Wu R (March 1974). "DNA sequence analysis: a general, simple and rapid method for sequencing large oligodeoxyribonucleotide fragments by mapping". Nucleic Acids Research. 1 (3): 331–53. doi:10.1093/nar/1.3.331. PMC 344020. PMID 10793670.
- ^ Hunt, Katie (17 February 2021). "World's oldest DNA sequenced from a mammoth that lived more than a million years ago". CNN. Retrieved 17 February 2021.
- ^ Callaway, Ewen (17 February 2021). "Million-year-old mammoth genomes shatter record for oldest ancient DNA – Permafrost-preserved teeth, up to 1.6 million years old, identify a new kind of mammoth in Siberia" (PDF). Nature. 590 (7847): 537–538. Bibcode:2021Natur.590..537C. doi:10.1038/d41586-021-00436-x. PMID 33597786. Retrieved 16 August 2025.
- ^ a b c Castro, Christina; Marine, Rachel; Ramos, Edward; Ng, Terry Fei Fan (2019). "The effect of variant interference on de novo assembly for viral deep sequencing". BMC Genomics. 21 (1): 421. bioRxiv 10.1101/815480. doi:10.1186/s12864-020-06801-w. PMC 7306937. PMID 32571214.
- ^ a b Wohl, Shirlee; Schaffner, Stephen F.; Sabeti, Pardis C. (2016). "Genomic Analysis of Viral Outbreaks". Annual Review of Virology. 3 (1): 173–195. doi:10.1146/annurev-virology-110615-035747. PMC 5210220. PMID 27501264.
- ^ Boycott, Kym M.; Vanstone, Megan R.; Bulman, Dennis E.; MacKenzie, Alex E. (October 2013). "Rare-disease genetics in the era of next-generation sequencing: discovery to translation". Nature Reviews Genetics. 14 (10): 681–691. doi:10.1038/nrg3555. PMID 23999272. S2CID 8496181.
- ^ Bean, Lora; Funke, Birgit; Carlston, Colleen M.; Gannon, Jennifer L.; Kantarci, Sibel; Krock, Bryan L.; Zhang, Shulin; Bayrak-Toydemir, Pinar (March 2020). "Diagnostic gene sequencing panels: from design to report—a technical standard of the American College of Medical Genetics and Genomics (ACMG)". Genetics in Medicine. 22 (3): 453–461. doi:10.1038/s41436-019-0666-z. ISSN 1098-3600. PMID 31732716.
- ^ Schleusener V, Köser CU, Beckert P, Niemann S, Feuerriegel S (2017). "Mycobacterium tuberculosis resistance prediction and lineage classification from genome sequencing: comparison of automated analysis tools". Sci Rep. 7 46327. Bibcode:2017NatSR...746327S. doi:10.1038/srep46327. PMC 7365310. PMID 28425484.
- ^ Mahé P, El Azami M, Barlas P, Tournoud M (2019). "A large scale evaluation of TBProfiler and Mykrobe for antibiotic resistance prediction in Mycobacterium tuberculosis". PeerJ. 7 e6857. doi:10.7717/peerj.6857. PMC 6500375. PMID 31106066.
- ^ Mykrobe predictor –Antibiotic resistance prediction for S. aureus and M. tuberculosis from whole genome sequence data
- ^ Bradley, Phelim; Gordon, N. Claire; Walker, Timothy M.; Dunn, Laura; Heys, Simon; Huang, Bill; Earle, Sarah; Pankhurst, Louise J.; Anson, Luke; de Cesare, Mariateresa; Piazza, Paolo; Votintseva, Antonina A.; Golubchik, Tanya; Wilson, Daniel J.; Wyllie, David H.; Diel, Roland; Niemann, Stefan; Feuerriegel, Silke; Kohl, Thomas A.; Ismail, Nazir; Omar, Shaheed V.; Smith, E. Grace; Buck, David; McVean, Gil; Walker, A. Sarah; Peto, Tim E. A.; Crook, Derrick W.; Iqbal, Zamin (21 December 2015). "Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis". Nature Communications. 6 (1) 10063. Bibcode:2015NatCo...610063B. doi:10.1038/ncomms10063. PMC 4703848. PMID 26686880.
- ^ "Michael Mosley vs the superbugs". Archived from the original on 24 November 2020. Retrieved 21 October 2019.
- ^ Mykrobe, Mykrobe-tools, 24 December 2022, retrieved 2 January 2023
- ^ Curtis C, Hereward J (29 August 2017). "From the crime scene to the courtroom: the journey of a DNA sample". The Conversation.
- ^ Moréra S, Larivière L, Kurzeck J, Aschke-Sonnenborn U, Freemont PS, Janin J, Rüger W (August 2001). "High resolution crystal structures of T4 phage beta-glucosyltransferase: induced fit and effect of substrate and metal binding". Journal of Molecular Biology. 311 (3): 569–77. doi:10.1006/jmbi.2001.4905. PMID 11493010.
- ^ Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, Gehrke C (April 1982). "Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells". Nucleic Acids Research. 10 (8): 2709–21. doi:10.1093/nar/10.8.2709. PMC 320645. PMID 7079182.
- ^ Ehrlich M, Wang RY (June 1981). "5-Methylcytosine in eukaryotic DNA". Science. 212 (4501): 1350–7. Bibcode:1981Sci...212.1350E. doi:10.1126/science.6262918. PMID 6262918.
- ^ Song CX, Clark TA, Lu XY, Kislyuk A, Dai Q, Turner SW, et al. (November 2011). "Sensitive and specific single-molecule sequencing of 5-hydroxymethylcytosine". Nature Methods. 9 (1): 75–7. doi:10.1038/nmeth.1779. PMC 3646335. PMID 22101853.
- ^ Czernecki, Dariusz; Bonhomme, Frédéric; Kaminski, Pierre-Alexandre; Delarue, Marc (5 August 2021). "Characterization of a triad of genes in cyanophage S-2L sufficient to replace adenine by 2-aminoadenine in bacterial DNA". Nature Communications. 12 (1): 4710. Bibcode:2021NatCo..12.4710C. doi:10.1038/s41467-021-25064-x. PMC 8342488. PMID 34354070. S2CID 233745192.
- ^ "Direct detection and sequencing of damaged DNA bases". PacBio. Retrieved 31 July 2024.
- ^ Watson JD, Crick FH (1953). "The structure of DNA". Cold Spring Harb. Symp. Quant. Biol. 18: 123–31. doi:10.1101/SQB.1953.018.01.020. PMID 13168976.
- ^ Marks, L. "The path to DNA sequencing: The life and work of Frederick Sanger". What is Biotechnology?. Retrieved 27 June 2023.
- ^ Min Jou W, Haegeman G, Ysebaert M, Fiers W (May 1972). "Nucleotide sequence of the gene coding for the bacteriophage MS2 coat protein". Nature. 237 (5350): 82–8. Bibcode:1972Natur.237...82J. doi:10.1038/237082a0. PMID 4555447. S2CID 4153893.
- ^ Fiers W, Contreras R, Duerinck F, Haegeman G, Iserentant D, Merregaert J, Min Jou W, Molemans F, Raeymaekers A, Van den Berghe A, Volckaert G, Ysebaert M (April 1976). "Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene". Nature. 260 (5551): 500–7. Bibcode:1976Natur.260..500F. doi:10.1038/260500a0. PMID 1264203. S2CID 4289674.
- ^ Ozsolak F, Milos PM (February 2011). "RNA sequencing: advances, challenges and opportunities". Nature Reviews Genetics. 12 (2): 87–98. doi:10.1038/nrg2934. PMC 3031867. PMID 21191423.
- ^ "Ray Wu Faculty Profile". Cornell University. Archived from the original on 4 March 2009.
- ^ Padmanabhan R, Jay E, Wu R (June 1974). "Chemical synthesis of a primer and its use in the sequence analysis of the lysozyme gene of bacteriophage T4". Proceedings of the National Academy of Sciences of the United States of America. 71 (6): 2510–4. Bibcode:1974PNAS...71.2510P. doi:10.1073/pnas.71.6.2510. PMC 388489. PMID 4526223.
- ^ Onaga LA (June 2014). "Ray Wu as Fifth Business: Demonstrating Collective Memory in the History of DNA Sequencing". Studies in the History and Philosophy of Science. Part C. 46: 1–14. doi:10.1016/j.shpsc.2013.12.006. PMID 24565976.
- ^ Wu R (1972). "Nucleotide sequence analysis of DNA". Nature New Biology. 236 (68): 198–200. doi:10.1038/newbio236198a0. PMID 4553110.
- ^ Padmanabhan R, Wu R (1972). "Nucleotide sequence analysis of DNA. IX. Use of oligonucleotides of defined sequence as primers in DNA sequence analysis". Biochem. Biophys. Res. Commun. 48 (5): 1295–302. Bibcode:1972BBRC...48.1295P. doi:10.1016/0006-291X(72)90852-2. PMID 4560009.
- ^ Wu R, Tu CD, Padmanabhan R (1973). "Nucleotide sequence analysis of DNA. XII. The chemical synthesis and sequence analysis of a dodecadeoxynucleotide which binds to the endolysin gene of bacteriophage lambda". Biochem. Biophys. Res. Commun. 55 (4): 1092–99. Bibcode:1973BBRC...55.1092R. doi:10.1016/S0006-291X(73)80007-5. PMID 4358929.
- ^ a b c Maxam AM, Gilbert W (February 1977). "A new method for sequencing DNA". Proc. Natl. Acad. Sci. USA. 74 (2): 560–64. Bibcode:1977PNAS...74..560M. doi:10.1073/pnas.74.2.560. PMC 392330. PMID 265521.
- ^ Gilbert, W. DNA sequencing and gene structure. Nobel lecture, 8 December 1980.
- ^ Gilbert W, Maxam A (December 1973). "The Nucleotide Sequence of the lac Operator". Proc. Natl. Acad. Sci. U.S.A. 70 (12): 3581–84. Bibcode:1973PNAS...70.3581G. doi:10.1073/pnas.70.12.3581. PMC 427284. PMID 4587255.
- ^ "Chapter 5: Investigating DNA". Chemistry. Retrieved 31 January 2025.
- ^ Sanger, F.; Coulson, A. R. (25 May 1975). "A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase". Journal of Molecular Biology. 94 (3): 441–448. doi:10.1016/0022-2836(75)90213-2. ISSN 0022-2836. PMID 1100841.
- ^ Cook-Deegan, Robert (1995). The gene wars: science, politics, and the human genome (1. publ. as a Norton paperback ed.). New York NY: Norton. ISBN 978-0-393-31399-4.
- ^ Johnson, Carolyn Y. (12 March 2015). "A physicist, biologist, Nobel laureate, CEO, and now, artist". The Boston Globe. Retrieved 3 February 2025.
- ^ a b c Heather, James M.; Chain, Benjamin (January 2016). "The sequence of sequencers: The history of sequencing DNA". Genomics. 107 (1): 1–8. doi:10.1016/j.ygeno.2015.11.003. PMC 4727787. PMID 26554401.
- ^ Deharvengt, Sophie J.; Petersen, Lauren M.; Jung, Hou-Sung; Tsongalis, Gregory J. (2020). "Nucleic acid analysis in the clinical laboratory". Contemporary Practice in Clinical Chemistry. pp. 215–234. doi:10.1016/B978-0-12-815499-1.00013-2. ISBN 978-0-12-815499-1.
- ^ Elsayed, Fadwa A.; Grolleman, Judith E.; Ragunathan, Abiramy; Buchanan, Daniel D.; van Wezel, Tom; de Voer, Richarda M.; Boot, Arnoud; Stojovska, Marija Staninova; Mahmood, Khalid; Clendenning, Mark; de Miranda, Noel; Dymerska, Dagmara; Egmond, Demi van; Gallinger, Steven; Georgeson, Peter; Hoogerbrugge, Nicoline; Hopper, John L.; Jansen, Erik A.M.; Jenkins, Mark A.; Joo, Jihoon E.; Kuiper, Roland P.; Ligtenberg, Marjolijn J.L.; Lubinski, Jan; Macrae, Finlay A.; Morreau, Hans; Newcomb, Polly; Nielsen, Maartje; Palles, Claire; Park, Daniel J.; Pope, Bernard J.; Rosty, Christophe; Ruiz Ponte, Clara; Schackert, Hans K.; Sijmons, Rolf H.; Tomlinson, Ian P.; Tops, Carli M.J.; Vreede, Lilian; Walker, Romy; Win, Aung K. (December 2020). "Monoallelic NTHL1 Loss-of-Function Variants and Risk of Polyposis and Colorectal Cancer". Gastroenterology. 159 (6): 2241–2243.e6. doi:10.1053/j.gastro.2020.08.042. hdl:2066/228713. PMC 7899696. PMID 32860789.
- ^ a b Sanger F, Nicklen S, Coulson AR (December 1977). "DNA sequencing with chain-terminating inhibitors". Proc. Natl. Acad. Sci. USA. 74 (12): 5463–77. Bibcode:1977PNAS...74.5463S. doi:10.1073/pnas.74.12.5463. PMC 431765. PMID 271968.
- ^ Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA, Hutchison CA, Slocombe PM, Smith M (February 1977). "Nucleotide sequence of bacteriophage phi X174 DNA". Nature. 265 (5596): 687–95. Bibcode:1977Natur.265..687S. doi:10.1038/265687a0. PMID 870828. S2CID 4206886.
- ^ Marks, L. "The next frontier: Human viruses". What is Biotechnology?. Retrieved 27 June 2023.
- ^ Beck S, Pohl FM (1984). "DNA sequencing with direct blotting electrophoresis". EMBO J. 3 (12): 2905–09. doi:10.1002/j.1460-2075.1984.tb02230.x. PMC 557787. PMID 6396083.
- ^ United States Patent 4,631,122 (1986)
- ^ Feldmann H, et al. (1994). "Complete DNA sequence of yeast chromosome II". EMBO J. 13 (24): 5795–809. doi:10.1002/j.1460-2075.1994.tb06923.x. PMC 395553. PMID 7813418.
- ^ Smith LM, Sanders JZ, Kaiser RJ, Hughes P, Dodd C, Connell CR, Heiner C, Kent SB, Hood LE (12 June 1986). "Fluorescence Detection in Automated DNA Sequence Analysis". Nature. 321 (6071): 674–79. Bibcode:1986Natur.321..674S. doi:10.1038/321674a0. PMID 3713851. S2CID 27800972.
- ^ Prober JM, Trainor GL, Dam RJ, Hobbs FW, Robertson CW, Zagursky RJ, Cocuzza AJ, Jensen MA, Baumeister K (16 October 1987). "A system for rapid DNA sequencing with fluorescent chain-terminating dideoxynucleotides". Science. 238 (4825): 336–41. Bibcode:1987Sci...238..336P. doi:10.1126/science.2443975. PMID 2443975.
- ^ Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF (June 1991). "Complementary DNA sequencing: expressed sequence tags and human genome project". Science. 252 (5013): 1651–56. Bibcode:1991Sci...252.1651A. doi:10.1126/science.2047873. PMID 2047873. S2CID 13436211.
- ^ Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM (July 1995). "Whole-genome random sequencing and assembly of Haemophilus influenzae Rd". Science. 269 (5223): 496–512. Bibcode:1995Sci...269..496F. doi:10.1126/science.7542800. PMID 7542800.
- ^ Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. (February 2001). "Initial sequencing and analysis of the human genome" (PDF). Nature. 409 (6822): 860–921. Bibcode:2001Natur.409..860L. doi:10.1038/35057062. PMID 11237011.
- ^ Venter JC, Adams MD, et al. (February 2001). "The sequence of the human genome". Science. 291 (5507): 1304–51. Bibcode:2001Sci...291.1304V. doi:10.1126/science.1058040. PMID 11181995.
- ^ "First complete sequence of a human genome". National Institutes of Health (NIH). 11 April 2022. Retrieved 6 February 2025.
- ^ "First complete sequence of a human genome". National Institutes of Health (NIH). 11 April 2022. Retrieved 6 February 2025.
- ^ Hartley, Gabrielle (31 March 2022). "The Human Genome Project pieced together only 92% of the DNA – now scientists have finally filled in the remaining 8%". The Conversation. Retrieved 6 February 2025.
- ^ Yang, Aimin; Zhang, Wei; Wang, Jiahao; Yang, Ke; Han, Yang; Zhang, Limin (2020). "Review on the Application of Machine Learning Algorithms in the Sequence Data Mining of DNA". Frontiers in Bioengineering and Biotechnology. 8 1032. doi:10.3389/fbioe.2020.01032. PMC 7498545. PMID 33015010.
- ^ Lyman DF, Bell A, Black A, Dingerdissen H, Cauley E, Gogate N, Liu D, Joseph A, Kahsay R, Crichton DJ, Mehta A, Mazumder R (19 September 2022). "Modeling and integration of N-glycan biomarkers in a comprehensive biomarker data model". Glycobiology. 32 (10): 855–870. doi:10.1093/glycob/cwac046. PMC 9487899. PMID 35925813. Retrieved 17 August 2025.
- ^ "Espacenet – Bibliographic data". worldwide.espacenet.com. Archived from the original on 10 January 2022. Retrieved 13 February 2015.
- ^ Ronaghi M, Karamohamed S, Pettersson B, Uhlén M, Nyrén P (1996). "Real-time DNA sequencing using detection of pyrophosphate release". Analytical Biochemistry. 242 (1): 84–89. doi:10.1006/abio.1996.0432. PMID 8923969.
- ^ a b Kawashima, Eric H.; Laurent Farinelli; Pascal Mayer (12 May 2005). "Patent: Method of nucleic acid amplification". Archived from the original on 22 February 2013. Retrieved 22 December 2012.
- ^ Ewing B, Green P (March 1998). "Base-calling of automated sequencer traces using phred. II. Error probabilities". Genome Res. 8 (3): 186–94. doi:10.1101/gr.8.3.186. PMID 9521922.
- ^ "Quality Scores for Next-Generation Sequencing" (PDF). Illumina. 31 October 2011. Retrieved 8 May 2018.
- ^ a b Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, Roth R, George D, Eletr S, Albrecht G, Vermaas E, Williams SR, Moon K, Burcham T, Pallas M, DuBridge RB, Kirchner J, Fearon K, Mao J, Corcoran K (2000). "Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays". Nature Biotechnology. 18 (6): 630–34. doi:10.1038/76469. PMID 10835600. S2CID 13884154.
- ^ "maxam gilbert sequencing". PubMed.
- ^ Sanger F, Coulson AR (May 1975). "A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase". J. Mol. Biol. 94 (3): 441–48. doi:10.1016/0022-2836(75)90213-2. PMID 1100841.
- ^ Wetterstrand, Kris. "DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP)". National Human Genome Research Institute. Retrieved 30 May 2013.
- ^ Nyren, P.; Pettersson, B.; Uhlen, M. (January 1993). "Solid Phase DNA Minisequencing by an Enzymatic Luminometric Inorganic Pyrophosphate Detection Assay". Analytical Biochemistry. 208 (1): 171–175. doi:10.1006/abio.1993.1024. PMID 8382019.
- ^ Ronaghi, Mostafa; Uhlén, Mathias; Nyrén, Pål (17 July 1998). "A Sequencing Method Based on Real-Time Pyrophosphate". Science. 281 (5375): 363–365. doi:10.1126/science.281.5375.363. PMID 9705713. S2CID 26331871.
- ^ Quail MA, Gu Y, Swerdlow H, Mayho M (2012). "Evaluation and optimisation of preparative semi-automated electrophoresis systems for Illumina library preparation". Electrophoresis. 33 (23): 3521–28. doi:10.1002/elps.201200128. PMID 23147856. S2CID 39818212.
- ^ Duhaime MB, Deng L, Poulos BT, Sullivan MB (2012). "Towards quantitative metagenomics of wild viruses and other ultra-low concentration DNA samples: a rigorous assessment and optimization of the linker amplification method". Environ. Microbiol. 14 (9): 2526–37. Bibcode:2012EnvMi..14.2526D. doi:10.1111/j.1462-2920.2012.02791.x. PMC 3466414. PMID 22713159.
- ^ Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE (2012). "Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species". PLOS ONE. 7 (5) e37135. Bibcode:2012PLoSO...737135P. doi:10.1371/journal.pone.0037135. PMC 3365034. PMID 22675423.
- ^ Williams R, Peisajovich SG, Miller OJ, Magdassi S, Tawfik DS, Griffiths AD (2006). "Amplification of complex gene libraries by emulsion PCR". Nature Methods. 3 (7): 545–50. doi:10.1038/nmeth896. PMID 16791213. S2CID 27459628.
- ^ a b Margulies M, Egholm M, et al. (September 2005). "Genome Sequencing in Open Microfabricated High Density Picoliter Reactors". Nature. 437 (7057): 376–80. Bibcode:2005Natur.437..376M. doi:10.1038/nature03959. PMC 1464427. PMID 16056220.
- ^ a b c Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM (9 September 2005). "Accurate Multiplex Polony Sequencing of an Evolved Bacterial Genome". Science. 309 (5741): 1728–32. Bibcode:2005Sci...309.1728S. doi:10.1126/science.1117389. PMID 16081699. S2CID 11405973.
- ^ "Applied Biosystems – File Not Found (404 Error)". 16 May 2008. Archived from the original on 16 May 2008.
- ^ Goodwin S, McPherson JD, McCombie WR (May 2016). "Coming of age: ten years of next-generation sequencing technologies". Nature Reviews Genetics. 17 (6): 333–51. doi:10.1038/nrg.2016.49. PMC 10373632. PMID 27184599. S2CID 8295541.
- ^ Staden R (11 June 1979). "A strategy of DNA sequencing employing computer programs". Nucleic Acids Research. 6 (7): 2601–10. doi:10.1093/nar/6.7.2601. PMC 327874. PMID 461197.
- ^ de Magalhães JP, Finch CE, Janssens G (2010). "Next-generation sequencing in aging research: emerging applications, problems, pitfalls and possible solutions". Ageing Research Reviews. 9 (3): 315–23. doi:10.1016/j.arr.2009.10.006. PMC 2878865. PMID 19900591.
- ^ Grada A (August 2013). "Next-generation sequencing: methodology and application". J Invest Dermatol. 133 (8): e11. doi:10.1038/jid.2013.248. PMID 23856935.
- ^ Hall N (May 2007). "Advanced sequencing technologies and their wider impact in microbiology". J. Exp. Biol. 210 (Pt 9): 1518–25. Bibcode:2007JExpB.210.1518H. doi:10.1242/jeb.001370. PMID 17449817.

- ^ Church GM (January 2006). "Genomes for all". Sci. Am. 294 (1): 46–54. Bibcode:2006SciAm.294a..46C. doi:10.1038/scientificamerican0106-46. PMID 16468433. S2CID 28769137.(subscription required)
- ^ a b c Schuster SC (January 2008). "Next-generation sequencing transforms today's biology". Nat. Methods. 5 (1): 16–18. doi:10.1038/nmeth1156. PMID 18165802. S2CID 1465786.
- ^ Kalb, Gilbert; Moxley, Robert (1992). Massively Parallel, Optical, and Neural Computing in the United States. IOS Press. ISBN 978-90-5199-097-3.[page needed]
- ^ ten Bosch JR, Grody WW (2008). "Keeping Up with the Next Generation". The Journal of Molecular Diagnostics. 10 (6): 484–92. doi:10.2353/jmoldx.2008.080027. PMC 2570630. PMID 18832462.

- ^ Tucker T, Marra M, Friedman JM (2009). "Massively Parallel Sequencing: The Next Big Thing in Genetic Medicine". The American Journal of Human Genetics. 85 (2): 142–54. doi:10.1016/j.ajhg.2009.06.022. PMC 2725244. PMID 19679224.

- ^ a b Straiton J, Free T, Sawyer A, Martin J (February 2019). "From Sanger sequencing to genome databases and beyond". BioTechniques. 66 (2). Future Science: 60–63. doi:10.2144/btn-2019-0011. PMID 30744413.
- ^ Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y (1 January 2012). "A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and illumina MiSeq sequencers". BMC Genomics. 13 (1): 341. doi:10.1186/1471-2164-13-341. PMC 3431227. PMID 22827831.

- ^ Liu L, Li Y, Li S, Hu N, He Y, Pong R, Lin D, Lu L, Law M (1 January 2012). "Comparison of Next-Generation Sequencing Systems". Journal of Biomedicine and Biotechnology. 2012 251364. doi:10.1155/2012/251364. PMC 3398667. PMID 22829749.

- ^ a b c "New Software, Polymerase for Sequel System Boost Throughput and Affordability – PacBio". 7 March 2018.
- ^ "After a Year of Testing, Two Early PacBio Customers Expect More Routine Use of RS Sequencer in 2012". GenomeWeb. 10 January 2012.(registration required)
- ^ "Pacific Biosciences Introduces New Chemistry With Longer Read Lengths to Detect Novel Features in DNA Sequence and Advance Genome Studies of Large Organisms" (Press release). 2013.
- ^ Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J (2013). "Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data". Nat. Methods. 10 (6): 563–69. doi:10.1038/nmeth.2474. PMID 23644548. S2CID 205421576.
- ^ a b "De novo bacterial genome assembly: a solved problem?". 5 July 2013.
- ^ Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, Paxinos EE, Sebra R, Chin CS, Iliopoulos D, Klammer A, Peluso P, Lee L, Kislyuk AO, Bullard J, Kasarskis A, Wang S, Eid J, Rank D, Redman JC, Steyert SR, Frimodt-Møller J, Struve C, Petersen AM, Krogfelt KA, Nataro JP, Schadt EE, Waldor MK (25 August 2011). "Origins of the Strain Causing an Outbreak of Hemolytic–Uremic Syndrome in Germany". N Engl J Med. 365 (8): 709–17. doi:10.1056/NEJMoa1106920. PMC 3168948. PMID 21793740.

- ^ Tran B, Brown AM, Bedard PL, Winquist E, Goss GD, Hotte SJ, Welch SA, Hirte HW, Zhang T, Stein LD, Ferretti V, Watt S, Jiao W, Ng K, Ghai S, Shaw P, Petrocelli T, Hudson TJ, Neel BG, Onetto N, Siu LL, McPherson JD, Kamel-Reid S, Dancey JE (1 January 2012). "Feasibility of real time next generation sequencing of cancer genes linked to drug response: Results from a clinical trial". Int. J. Cancer. 132 (7): 1547–55. doi:10.1002/ijc.27817. PMID 22948899. S2CID 72705.(subscription required)
- ^ Murray IA, Clark TA, Morgan RD, Boitano M, Anton BP, Luong K, Fomenkov A, Turner SW, Korlach J, Roberts RJ (2 October 2012). "The methylomes of six bacteria". Nucleic Acids Research. 40 (22): 11450–62. doi:10.1093/nar/gks891. PMC 3526280. PMID 23034806.
- ^ "Ion 520 & Ion 530 ExT Kit-Chef – Thermo Fisher Scientific". thermofisher.com.
- ^ "Raw accuracy". Archived from the original on 30 March 2018. Retrieved 29 March 2018.
- ^ van Vliet AH (1 January 2010). "Next generation sequencing of microbial transcriptomes: challenges and opportunities". FEMS Microbiology Letters. 302 (1): 1–7. doi:10.1111/j.1574-6968.2009.01767.x. PMID 19735299.

- ^ "BGI and MGISEQ". en.mgitech.cn. Archived from the original on 7 July 2018. Retrieved 5 July 2018.
- ^ a b Huang YF, Chen SC, Chiang YS, Chen TH, Chiu KP (2012). "Palindromic sequence impedes sequencing-by-ligation mechanism". BMC Systems Biology. 6 (Suppl 2) S10. doi:10.1186/1752-0509-6-S2-S10. PMC 3521181. PMID 23281822.
- ^ Loose, Matthew; Rakyan, Vardhman; Holmes, Nadine; Payne, Alexander (3 May 2018). "Whale watching with BulkVis: A graphical viewer for Oxford Nanopore bulk fast5 files". bioRxiv 10.1101/312256.
- ^ "PacBio Sales Start to Pick Up as Company Delivers on Product Enhancements". 12 February 2013.
- ^ "Bio-IT World". bio-itworld.com. Archived from the original on 29 July 2020. Retrieved 16 November 2015.
- ^ "PacBio Launches Higher-Throughput, Lower-Cost Single-Molecule Sequencing System". October 2015.
- ^ Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H (April 2009). "Continuous base identification for single-molecule nanopore DNA sequencing". Nature Nanotechnology. 4 (4): 265–70. Bibcode:2009NatNa...4..265C. doi:10.1038/nnano.2009.12. PMID 19350039.
- ^ a b dela Torre R, Larkin J, Singer A, Meller A (2012). "Fabrication and characterization of solid-state nanopore arrays for high-throughput DNA sequencing". Nanotechnology. 23 (38) 385308. Bibcode:2012Nanot..23L5308D. doi:10.1088/0957-4484/23/38/385308. PMC 3557807. PMID 22948520.
- ^ a b Pathak B, Lofas H, Prasongkit J, Grigoriev A, Ahuja R, Scheicher RH (2012). "Double-functionalized nanopore-embedded gold electrodes for rapid DNA sequencing". Applied Physics Letters. 100 (2) 023701. Bibcode:2012ApPhL.100b3701P. doi:10.1063/1.3673335.
- ^ Korlach J, Marks PJ, Cicero RL, Gray JJ, Murphy DL, Roitman DB, Pham TT, Otto GA, Foquet M, Turner SW (2008). "Selective aluminum passivation for targeted immobilization of single DNA polymerase molecules in zero-mode waveguide nanostructures". Proceedings of the National Academy of Sciences. 105 (4): 1176–81. Bibcode:2008PNAS..105.1176K. doi:10.1073/pnas.0710982105. PMC 2234111. PMID 18216253.
- ^ Bentley DR, Balasubramanian S, et al. (2008). "Accurate whole human genome sequencing using reversible terminator chemistry". Nature. 456 (7218): 53–59. Bibcode:2008Natur.456...53B. doi:10.1038/nature07517. PMC 2581791. PMID 18987734.
- ^ Canard B, Sarfati S (13 October 1994), Novel derivatives usable for the sequencing of nucleic acids, retrieved 9 March 2016
- ^ Canard B, Sarfati RS (October 1994). "DNA polymerase fluorescent substrates with reversible 3'-tags". Gene. 148 (1): 1–6. doi:10.1016/0378-1119(94)90226-7. PMID 7523248.
- ^ Mardis ER (2008). "Next-generation DNA sequencing methods". Annu Rev Genom Hum Genet. 9: 387–402. doi:10.1146/annurev.genom.9.081307.164359. PMID 18576944.
- ^ a b c Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, et al. (January 2010). "Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays". Science. 327 (5961): 78–81. Bibcode:2010Sci...327...78D. doi:10.1126/science.1181498. PMID 19892942. S2CID 17309571.
- ^ brandonvd. "About Us – Complete Genomics". Complete Genomics. Retrieved 2 July 2018.
- ^ a b Huang J, Liang X, Xuan Y, Geng C, Li Y, Lu H, et al. (May 2017). "A reference human genome dataset of the BGISEQ-500 sequencer". GigaScience. 6 (5) gix024: 1–9. doi:10.1093/gigascience/gix024. PMC 5467036. PMID 28379488.
- ^ Bornman DM, Hester ME, Schuetter JM, Kasoji MD, Minard-Smith A, Barden CA, Nelson SC, Godbold GD, Baker CH, Yang B, Walther JE, Tornes IE, Yan PS, Rodriguez B, Bundschuh R, Dickens ML, Young BA, Faith SA (1 April 2012). "Short-read, high-throughput sequencing technology for STR genotyping" (PDF). Biotech Rapid Dispatches. 2012: 1–6. PMC 4301848. PMID 25621315. Archived from the original (PDF) on 8 August 2017. Retrieved 16 August 2025.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Li Q, Zhao X, Zhang W, Wang L, Wang J, Xu D, Mei Z, Liu Q, Du S, Li Z, Liang X, Wang X, Wei H, Liu P, Zou J, Shen H, Chen A, Drmanac S, Liu JS, Li L, Jiang H, Zhang Y, Wang J, Yang H, Xu X, Drmanac R, Jiang Y (13 March 2019). "Reliable multiplex sequencing with rare index mis-assignment on DNB-based NGS platform". BMC Genomics. 20 (1) 215. doi:10.1186/s12864-019-5569-5. PMC 6416933. PMID 30866797.
- ^ Holt RA, Jones SJ (June 2008). "The new paradigm of flow cell sequencing" (PDF). Genome Res. 18 (6): 839–846. doi:10.1101/gr.073262.107. PMID 18519653. Retrieved 16 August 2025.
- ^ Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, Sidow A, Fire A, Johnson SM (July 2008). "A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning". Genome Res. 18 (7): 1051–63. doi:10.1101/gr.076463.108. PMC 2493394. PMID 18477713.
- ^ Rusk N (2011). "Torrents of sequence". Nat Methods. 8 (1): 44. doi:10.1038/nmeth.f.330. S2CID 41040192.
- ^ a b Drmanac R, Sparks AB, et al. (2010). "Human Genome Sequencing Using Unchained Base Reads in Self-Assembling DNA Nanoarrays". Science. 327 (5961): 78–81. Bibcode:2010Sci...327...78D. doi:10.1126/science.1181498. PMID 19892942. S2CID 17309571.
- ^ Porreca GJ (2010). "Genome Sequencing on Nanoballs". Nature Biotechnology. 28 (1): 43–44. doi:10.1038/nbt0110-43. PMID 20062041. S2CID 54557996.
- ^ "HeliScope Gene Sequencing / Genetic Analyzer System: Helicos BioSciences". 2 November 2009. Archived from the original on 2 November 2009.
- ^ Thompson JF, Steinmann KE (October 2010). "Single molecule sequencing with a HeliScope genetic analysis system". Current Protocols in Molecular Biology. Chapter 7: Unit7.10. doi:10.1002/0471142727.mb0710s92. PMC 2954431. PMID 20890904.
- ^ "tSMS SeqLL Technical Explanation". SeqLL. Archived from the original on 8 August 2014. Retrieved 9 August 2015.
- ^ Sara El-Metwally; Osama M. Ouda; Mohamed Helmy (2014). "New Horizons in Next-Generation Sequencing". Next Generation Sequencing Technologies and Challenges in Sequence Assembly. SpringerBriefs in Systems Biology. Vol. 7. Next Generation Sequencing Technologies and Challenges in Sequence Assembly, Springer Briefs in Systems Biology Volume 7. pp. 51–59. doi:10.1007/978-1-4939-0715-1_6. ISBN 978-1-4939-0714-4.
- ^ a b Fair RB, Khlystov A, Tailor TD, Ivanov V, Evans RD, Srinivasan V, Pamula VK, Pollack MG, Griffin PB, Zhou J (January 2007). "Chemical and Biological Applications of Digital-Microfluidic Devices". IEEE Design & Test of Computers. 24 (1): 10–24. Bibcode:2007IDTC...24...10F. CiteSeerX 10.1.1.559.1440. doi:10.1109/MDT.2007.8. hdl:10161/6987. S2CID 10122940.
- ^ a b Boles DJ, Benton JL, Siew GJ, Levy MH, Thwar PK, Sandahl MA, et al. (November 2011). "Droplet-based pyrosequencing using digital microfluidics". Analytical Chemistry. 83 (22): 8439–47. doi:10.1021/ac201416j. PMC 3690483. PMID 21932784.
- ^ Zilionis R, Nainys J, Veres A, Savova V, Zemmour D, Klein AM, Mazutis L (January 2017). "Single-cell barcoding and sequencing using droplet microfluidics". Nature Protocols. 12 (1): 44–73. doi:10.1038/nprot.2016.154. PMID 27929523. S2CID 767782.
- ^ "The Harvard Nanopore Group". Mcb.harvard.edu. Archived from the original on 21 February 2002. Retrieved 15 November 2009.
- ^ "Nanopore Sequencing Could Slash DNA Analysis Costs".
- ^ US patent 20060029957, ZS Genetics, "Systems and methods of analyzing nucleic acid polymers and related components", issued 14 July 2005
- ^ Xu M, Fujita D, Hanagata N (December 2009). "Perspectives and challenges of emerging single-molecule DNA sequencing technologies". Small. 5 (23): 2638–49. Bibcode:2009Small...5.2638X. doi:10.1002/smll.200900976. PMID 19904762.
- ^ Schadt EE, Turner S, Kasarskis A (2010). "A window into third-generation sequencing". Human Molecular Genetics. 19 (R2): R227–40. doi:10.1093/hmg/ddq416. PMID 20858600.
- ^ Xu M, Endres RG, Arakawa Y (2007). "The electronic properties of DNA bases". Small. 3 (9): 1539–43. Bibcode:2007Small...3.1539X. doi:10.1002/smll.200600732. PMID 17786897.
- ^ Di Ventra M (2013). "Fast DNA sequencing by electrical means inches closer". Nanotechnology. 24 (34) 342501. Bibcode:2013Nanot..24H2501D. doi:10.1088/0957-4484/24/34/342501. PMID 23899780. S2CID 140101884.
- ^ Ohshiro T, Matsubara K, Tsutsui M, Furuhashi M, Taniguchi M, Kawai T (2012). "Single-molecule electrical random resequencing of DNA and RNA". Sci Rep. 2 501. Bibcode:2012NatSR...2..501O. doi:10.1038/srep00501. PMC 3392642. PMID 22787559.
- ^ Hanna GJ, Johnson VA, Kuritzkes DR, Richman DD, Martinez-Picado J, Sutton L, Hazelwood JD, D'Aquila RT (1 July 2000). "Comparison of Sequencing by Hybridization and Cycle Sequencing for Genotyping of Human Immunodeficiency Virus Type 1 Reverse Transcriptase". J. Clin. Microbiol. 38 (7): 2715–21. doi:10.1128/JCM.38.7.2715-2721.2000. PMC 87006. PMID 10878069.
- ^ a b Morey M, Fernández-Marmiesse A, Castiñeiras D, Fraga JM, Couce ML, Cocho JA (2013). "A glimpse into past, present, and future DNA sequencing". Molecular Genetics and Metabolism. 110 (1–2): 3–24. doi:10.1016/j.ymgme.2013.04.024. hdl:20.500.11940/2036. PMID 23742747.
- ^ Qin Y, Schneider TM, Brenner MP (2012). Gibas C (ed.). "Sequencing by Hybridization of Long Targets". PLOS ONE. 7 (5) e35819. Bibcode:2012PLoSO...735819Q. doi:10.1371/journal.pone.0035819. PMC 3344849. PMID 22574124.
- ^ Edwards JR, Ruparel H, Ju J (2005). "Mass-spectrometry DNA sequencing". Mutation Research. 573 (1–2): 3–12. Bibcode:2005MRFMM.573....3E. doi:10.1016/j.mrfmmm.2004.07.021. PMID 15829234.
- ^ Hall TA, Budowle B, Jiang Y, Blyn L, Eshoo M, Sannes-Lowery KA, Sampath R, Drader JJ, Hannis JC, Harrell P, Samant V, White N, Ecker DJ, Hofstadler SA (2005). "Base composition analysis of human mitochondrial DNA using electrospray ionization mass spectrometry: A novel tool for the identification and differentiation of humans". Analytical Biochemistry. 344 (1): 53–69. doi:10.1016/j.ab.2005.05.028. PMID 16054106.
- ^ Howard R, Encheva V, Thomson J, Bache K, Chan YT, Cowen S, Debenham P, Dixon A, Krause JU, Krishan E, Moore D, Moore V, Ojo M, Rodrigues S, Stokes P, Walker J, Zimmermann W, Barallon R (15 June 2011). "Comparative analysis of human mitochondrial DNA from World War I bone samples by DNA sequencing and ESI-TOF mass spectrometry". Forensic Science International: Genetics. 7 (1): 1–9. doi:10.1016/j.fsigen.2011.05.009. PMID 21683667.
- ^ Monforte JA, Becker CH (1 March 1997). "High-throughput DNA analysis by time-of-flight mass spectrometry". Nature Medicine. 3 (3): 360–62. doi:10.1038/nm0397-360. PMID 9055869. S2CID 28386145.
- ^ Beres SB, Carroll RK, Shea PR, Sitkiewicz I, Martinez-Gutierrez JC, Low DE, McGeer A, Willey BM, Green K, Tyrrell GJ, Goldman TD, Feldgarden M, Birren BW, Fofanov Y, Boos J, Wheaton WD, Honisch C, Musser JM (8 February 2010). "Molecular complexity of successive bacterial epidemics deconvoluted by comparative pathogenomics". Proceedings of the National Academy of Sciences. 107 (9): 4371–76. Bibcode:2010PNAS..107.4371B. doi:10.1073/pnas.0911295107. PMC 2840111. PMID 20142485.
- ^ Kan CW, Fredlake CP, Doherty EA, Barron AE (1 November 2004). "DNA sequencing and genotyping in miniaturized electrophoresis systems". Electrophoresis. 25 (21–22): 3564–88. doi:10.1002/elps.200406161. PMID 15565709. S2CID 4851728.
- ^ Chen YJ, Roller EE, Huang X (2010). "DNA sequencing by denaturation: experimental proof of concept with an integrated fluidic device". Lab on a Chip. 10 (9): 1153–59. doi:10.1039/b921417h. PMC 2881221. PMID 20390134.
- ^ Bell DC, Thomas WK, Murtagh KM, Dionne CA, Graham AC, Anderson JE, Glover WR (9 October 2012). "DNA Base Identification by Electron Microscopy". Microscopy and Microanalysis. 18 (5): 1049–53. Bibcode:2012MiMic..18.1049B. doi:10.1017/S1431927612012615. PMID 23046798. S2CID 25713635.
- ^ a b Pareek CS, Smoczynski R, Tretyn A (November 2011). "Sequencing technologies and genome sequencing". Journal of Applied Genetics. 52 (4): 413–35. doi:10.1007/s13353-011-0057-x. PMC 3189340. PMID 21698376.
- ^ Fujimori S, Hirai N, Ohashi H, Masuoka K, Nishikimi A, Fukui Y, Washio T, Oshikubo T, Yamashita T, Miyamoto-Sato E (2012). "Next-generation sequencing coupled with a cell-free display technology for high-throughput production of reliable interactome data". Scientific Reports. 2 691. Bibcode:2012NatSR...2..691F. doi:10.1038/srep00691. PMC 3466446. PMID 23056904.
- ^ "2022 Sequencing Market Share – Same as It Ever Was (For Now)". 25 June 2023.
- ^ Harbers M (2008). "The Current Status of cDNA Cloning". Genomics. 91 (3): 232–42. doi:10.1016/j.ygeno.2007.11.004. PMID 18222633.
- ^ Alberti A, Belser C, Engelen S, Bertrand L, Orvain C, Brinas L, Cruaud C, et al. (2014). "Comparison of Library Preparation Methods Reveals Their Impact on Interpretation of Metatranscriptomic Data". BMC Genomics. 15 (1): 912–12. doi:10.1186/1471-2164-15-912. PMC 4213505. PMID 25331572.
- ^ "Scalable Nucleic Acid Quality Assessments for Illumina Next-Generation Sequencing Library Prep" (PDF). Retrieved 27 December 2017.
- ^ "Archon Genomics XPRIZE". Archon Genomics XPRIZE. Archived from the original on 17 June 2013. Retrieved 9 August 2007.
- ^ "Grant Information". National Human Genome Research Institute (NHGRI).
- ^ Severin J, Lizio M, Harshbarger J, Kawaji H, Daub CO, Hayashizaki Y, Bertin N, Forrest AR (2014). "Interactive visualization and analysis of large-scale sequencing datasets using ZENBU". Nat. Biotechnol. 32 (3): 217–19. doi:10.1038/nbt.2840. PMID 24727769. S2CID 26575621.
- ^ Shmilovici A, Ben-Gal I (2007). "Using a VOM model for reconstructing potential coding regions in EST sequences" (PDF). Computational Statistics. 22 (1): 49–69. doi:10.1007/s00180-007-0021-8. S2CID 2737235. Archived from the original (PDF) on 31 May 2020. Retrieved 10 January 2014.
- ^ Del Fabbro C, Scalabrin S, Morgante M, Giorgi FM (2013). "An Extensive Evaluation of Read Trimming Effects on Illumina NGS Data Analysis". PLOS ONE. 8 (12) e85024. Bibcode:2013PLoSO...885024D. doi:10.1371/journal.pone.0085024. PMC 3871669. PMID 24376861.
- ^ Martin, Marcel (2 May 2011). "Cutadapt removes adapter sequences from high-throughput sequencing reads". EMBnet.journal. 17 (1): 10. doi:10.14806/ej.17.1.200.
- ^ Smeds L, Künstner A (19 October 2011). "ConDeTri--a content dependent read trimmer for Illumina data". PLOS ONE. 6 (10) e26314. Bibcode:2011PLoSO...626314S. doi:10.1371/journal.pone.0026314. PMC 3198461. PMID 22039460.
- ^ Prezza N, Del Fabbro C, Vezzi F, De Paoli E, Policriti A (2012). "Erne-Bs5". Proceedings of the ACM Conference on Bioinformatics, Computational Biology and Biomedicine. Vol. 12. pp. 12–19. doi:10.1145/2382936.2382938. ISBN 978-1-4503-1670-5. S2CID 5673753.
- ^ Schmieder R, Edwards R (March 2011). "Quality control and preprocessing of metagenomic datasets". Bioinformatics. 27 (6): 863–4. doi:10.1093/bioinformatics/btr026. PMC 3051327. PMID 21278185.
- ^ Bolger AM, Lohse M, Usadel B (August 2014). "Trimmomatic: a flexible trimmer for Illumina sequence data". Bioinformatics. 30 (15): 2114–20. doi:10.1093/bioinformatics/btu170. PMC 4103590. PMID 24695404.
- ^ Cox MP, Peterson DA, Biggs PJ (September 2010). "SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data". BMC Bioinformatics. 11 (1) 485. doi:10.1186/1471-2105-11-485. PMC 2956736. PMID 20875133.
- ^ Murray TH (January 1991). "Ethical issues in human genome research". FASEB Journal. 5 (1): 55–60. doi:10.1096/fasebj.5.1.1825074. PMID 1825074. S2CID 20009748.
- ^ a b c Robertson JA (August 2003). "The $1000 genome: ethical and legal issues in whole genome sequencing of individuals". The American Journal of Bioethics. 3 (3): W–IF1. doi:10.1162/152651603322874762. PMID 14735880. S2CID 15357657. Retrieved 20 May 2015.
- ^ a b Henderson, Mark (9 September 2013). "Human genome sequencing: the real ethical dilemmas". The Guardian. Retrieved 20 May 2015.
- ^ Harmon, Amy (24 February 2008). "Insurance Fears Lead Many to Shun DNA Tests". The New York Times. Retrieved 20 May 2015.
- ^ Statement of Administration policy, Executive Office of the President, Office of Management and Budget, 27 April 2007
- ^ National Human Genome Research Institute (21 May 2008). "President Bush Signs the Genetic Information Nondiscrimination Act of 2008". Retrieved 17 February 2014.
- ^ Baker, Monya (11 October 2012). "US ethics panel reports on DNA sequencing and privacy". Nature News Blog.
- ^ "Privacy and Progress in Whole Genome Sequencing" (PDF). Presidential Commission for the Study of Bioethical Issues. Archived from the original (PDF) on 12 June 2015. Retrieved 20 May 2015.
- ^ Hartnett, Kevin (12 May 2013). "The DNA in your garbage: up for grabs". The Boston Globe. Retrieved 2 January 2023.
- ^ Goldenberg AJ, Sharp RR (February 2012). "The ethical hazards and programmatic challenges of genomic newborn screening". JAMA. 307 (5): 461–2. doi:10.1001/jama.2012.68. PMC 3868436. PMID 22298675.
- ^ Hughes, Virginia (7 January 2013). "It's Time To Stop Obsessing About the Dangers of Genetic Information". Slate Magazine. Retrieved 22 May 2015.
- ^ a b Bloss CS, Schork NJ, Topol EJ (February 2011). "Effect of direct-to-consumer genomewide profiling to assess disease risk". The New England Journal of Medicine. 364 (6): 524–34. doi:10.1056/NEJMoa1011893. PMC 3786730. PMID 21226570.
- ^ Rochman, Bonnie (25 October 2012). "What Your Doctor Isn't Telling You About Your DNA". Time. Retrieved 22 May 2015.
- ^ Sajeer P, Muhammad (4 May 2023). "Disruptive technology: Exploring the ethical, legal, political, and societal implications of nanopore sequencing technology". EMBO Reports. 24 (5) e56619. doi:10.15252/embr.202256619. PMC 10157308. PMID 36988424. S2CID 257803254.
External links
[edit]DNA sequencing
View on GrokipediaFundamentals
Nucleotide Composition and DNA Structure
DNA, or deoxyribonucleic acid, is a nucleic acid polymer composed of repeating nucleotide monomers linked by phosphodiester bonds. Each nucleotide consists of three components: a 2'-deoxyribose sugar molecule, a phosphate group attached to the 5' carbon of the sugar, and one of four nitrogenous bases—adenine (A), guanine (G), cytosine (C), or thymine (T)—bound to the 1' carbon via a glycosidic bond.[13][14] Adenine and guanine belong to the purine class, featuring a fused double-ring structure, whereas cytosine and thymine are pyrimidines with a single-ring structure.[15] The sugar-phosphate backbone forms through covalent linkages between the 3' hydroxyl of one deoxyribose and the phosphate of the adjacent nucleotide, creating directional polarity with distinct 5' and 3' ends.[14] This backbone provides structural stability, while the sequence of bases encodes genetic information. In the canonical B-form double helix, as elucidated by James Watson and Francis Crick in 1953 based on X-ray diffraction data from Rosalind Franklin and Maurice Wilkins, two antiparallel DNA strands coil around a common axis with approximately 10.5 base pairs per helical turn and a pitch of 3.4 nanometers.[16][17] The hydrophobic bases stack inward via van der Waals interactions, stabilized by hydrogen bonding between complementary pairs: adenine-thymine (two hydrogen bonds) and guanine-cytosine (three hydrogen bonds), ensuring specificity in base pairing (A pairs exclusively with T, G with C).[18] This antiparallel orientation— one strand running 5' to 3', the other 3' to 5'—facilitates replication and transcription processes. The major and minor grooves in the helix expose edges of the bases, allowing proteins to recognize specific sequences without unwinding the structure.[14] Nucleotide composition varies across genomes, often quantified by GC content (the percentage of guanine and cytosine bases), which influences DNA stability, melting temperature, and evolutionary patterns; for instance, thermophilic organisms exhibit higher GC content for thermal resilience.[14] In the context of sequencing, the linear order of these four bases along the strand constitutes the primary data output, as methods exploit base-specific chemical or enzymatic properties to infer this sequence.[19] The double-helical architecture necessitates denaturation or strand separation in many sequencing protocols to access individual strands for base readout.[20]Core Principles of Sequencing Reactions
DNA sequencing reactions generate populations of polynucleotide fragments whose lengths correspond precisely to the positions of nucleotides in the target DNA sequence, enabling sequence determination through subsequent size-based separation, typically via electrophoresis or capillary methods. This fragment ladder approach relies on either enzymatic synthesis or chemical degradation to produce terminations at each base position, with detection historically via radiolabeling and more recently through fluorescence or other signals.[21][22] In enzymatic sequencing reactions, DNA-dependent DNA polymerase catalyzes the template-directed polymerization of deoxynucleotide triphosphates (dNTPs)—dATP, dCTP, dGTP, and dTTP—onto a primer annealed to denatured, single-stranded template DNA, forming phosphodiester bonds via the 3'-hydroxyl group of the growing chain attacking the alpha-phosphate of incoming dNTPs.[21] To create sequence-specific chain terminations, reactions incorporate low ratios of dideoxynucleotide triphosphates (ddNTPs), analogs lacking the 3'-OH group; when a ddNTP base-pairs with its complementary template base, polymerase incorporates it but cannot extend further, yielding fragments ending at every occurrence of that base across multiple template molecules.[21] Each of the four ddNTPs (ddATP, ddCTP, ddGTP, ddTTP) is used in separate reactions or color-coded in multiplex formats, with incorporation fidelity depending on polymerase selectivity and reaction conditions like temperature and buffer composition.[22] Chemical sequencing reactions, in contrast, exploit base-specific reactivity to modify phosphodiester backbones without enzymatic synthesis. Reagents such as dimethyl sulfate alkylate guanines, piperidine cleaves at modified sites, hydrazine reacts with pyrimidines (thymine/cytosine), and formic acid depurinates adenines/guanine, producing alkali-labile breaks that generate 5'-labeled fragments terminating at targeted bases after piperidine treatment and denaturation.[21] These methods require end-labeling of DNA (e.g., with 32P) for detection and yield partial digests calibrated to average one cleavage per molecule, ensuring a complete set of fragments from the labeled end to each modifiable base.[21] Both paradigms depend on stochastic termination across billions of template copies to populate all positions statistically, with reaction efficiency influenced by factors like template secondary structure, base composition biases (e.g., GC-rich regions resisting denaturation), and reagent purity; incomplete reactions or non-specific cleavages can introduce artifacts resolved by running multiple lanes or replicates.[22] Modern variants extend these principles, such as reversible terminator ddNTPs in sequencing-by-synthesis, where blocked 3'-OH groups are cleaved post-detection to allow iterative extension.[23]Historical Development
Pre-1970s Foundations: DNA Discovery and Early Enzymology
In 1869, Swiss biochemist Friedrich Miescher isolated a novel phosphorus-rich acidic substance, termed nuclein, from the nuclei of leukocytes obtained from surgical pus bandages; this material was later recognized as deoxyribonucleic acid (DNA).[24] Miescher's extraction involved treating cells with pepsin to remove proteins and alkali to precipitate the nuclein, establishing DNA as a distinct cellular component separate from proteins.[25] Subsequent work by Phoebus Levene in the early 20th century identified DNA's building blocks as nucleotides—phosphate, deoxyribose, and four bases (adenine, thymine, guanine, cytosine)—though Levene erroneously proposed a repetitive tetranucleotide structure, hindering recognition of DNA's informational potential.[26] The identification of DNA as the genetic material emerged from transformation experiments building on Frederick Griffith's 1928 observation that heat-killed virulent pneumococci could transfer virulence to non-virulent strains in mice. In 1944, Oswald Avery, Colin MacLeod, and Maclyn McCarty demonstrated that purified DNA from virulent Streptococcus pneumoniae type III-S transformed non-virulent type II-R bacteria into stable virulent forms, resistant to protein-digesting enzymes but sensitive to DNase; this provided the first rigorous evidence that DNA, not protein, carries hereditary information.[27] Confirmation came in 1952 via Alfred Hershey and Martha Chase's bacteriophage T2 experiments, where radioactively labeled DNA (with phosphorus-32) entered Escherichia coli cells during infection and produced progeny phages, while labeled protein coats (sulfur-35) remained outside, definitively establishing DNA as the heritable substance over protein.[28] The double-helical structure of DNA, proposed by James Watson and Francis Crick in 1953, revealed its capacity to store sequence-specific genetic information through complementary base pairing (adenine-thymine, guanine-cytosine), enabling precise replication and laying the conceptual groundwork for sequencing as a means to decode that sequence. This model integrated X-ray diffraction data from Rosalind Franklin and Maurice Wilkins, showing antiparallel strands twisted into a right-handed helix with 10 base pairs per turn, and implied enzymatic mechanisms for unwinding, copying, and repair.[29] Early enzymology advanced these foundations through the 1956 isolation of DNA polymerase by Arthur Kornberg, who demonstrated its template-directed synthesis of DNA from deoxynucleoside triphosphates in E. coli extracts, requiring a primer and fidelity via base complementarity—earning Kornberg the 1959 Nobel Prize in Physiology or Medicine shared with Severo Ochoa for RNA polymerase.[30] This enzyme's characterization illuminated semi-conservative replication, confirmed by Matthew Meselson and Franklin Stahl's 1958 density-gradient experiments using nitrogen isotopes, and enabled initial in vitro manipulations like end-labeling DNA strands, precursors to enzymatic sequencing approaches. By the late 1960s, identification of exonucleases and ligases further supported controlled DNA degradation and joining, though full replication systems revealed complexities like multiple polymerases (e.g., Pol II and III discovered circa 1970), underscoring DNA's enzymatic vulnerability and manipulability essential for later sequencing innovations.[31]1970s Breakthroughs: Chemical and Enzymatic Methods
In 1977, two pivotal DNA sequencing techniques emerged, enabling the routine determination of nucleotide sequences for the first time and transforming molecular biology research. The chemical degradation method, developed by Allan Maxam and Walter Gilbert at Harvard University, relied on base-specific chemical cleavage of radioactively end-labeled DNA fragments, followed by size separation via polyacrylamide gel electrophoresis to generate readable ladders of fragments corresponding to each nucleotide position.[32] This approach used dimethyl sulfate for guanine, formic acid for adenine plus guanine, hydrazine for cytosine plus thymine, and piperidine for cleavage, producing partial digests that revealed the sequence when resolved on denaturing gels.[33] Independently, Frederick Sanger and his team at the MRC Laboratory of Molecular Biology in Cambridge introduced the enzymatic chain-termination method later that year, employing DNA polymerase I to synthesize complementary strands from a single-stranded template in the presence of normal deoxynucleotides (dNTPs) and low concentrations of chain-terminating dideoxynucleotides (ddNTPs), each specific to one base (ddATP, ddGTP, ddCTP, ddTTP).[34] The resulting fragments, terminated randomly at each occurrence of the corresponding base, were separated by gel electrophoresis, allowing sequence readout from the positions of bands in parallel lanes for each ddNTP. This method built on Sanger's earlier "plus and minus" technique from 1975 but offered greater efficiency and accuracy for longer reads, up to 200-400 bases.[34] Both techniques represented breakthroughs over prior laborious approaches like two-dimensional chromatography, which were limited to short oligonucleotides of 10-20 bases.[3] The Maxam-Gilbert method's chemical basis avoided enzymatic biases but required hazardous reagents and precise control of partial reactions, while Sanger's enzymatic approach was more amenable to automation and cloning-based template preparation using M13 vectors in subsequent refinements.[35] Together, they enabled the first complete sequencing of a DNA genome, the 5,386-base bacteriophage φX174 by Sanger's group in 1977, demonstrating feasibility for viral and eventually eukaryotic gene analysis.[34] These 1970s innovations laid the empirical foundation for genomics, with Sanger's method predominating due to its scalability and lower toxicity, though both coexisted into the 1980s.[3]1980s-1990s: Genome-Scale Projects and Automation
In the 1980s, automation of Sanger chain-termination sequencing addressed the labor-intensive limitations of manual radioactive gel-based methods by introducing fluorescent dye-labeled dideoxynucleotides and laser detection systems. Researchers at the California Institute of Technology, including Leroy Hood and Lloyd Smith, developed prototype instruments that eliminated the need for radioisotopes and manual band interpretation, enabling four-color detection in a single lane.[36][37] Applied Biosystems commercialized the first such device, the ABI 370A, in 1986, utilizing slab polyacrylamide gel electrophoresis to process up to 48 samples simultaneously and achieve read lengths of 300-500 bases per run.[38][39] These innovations increased throughput from hundreds to thousands of bases per day per instrument, reducing costs and errors while scaling capacity for larger datasets.[40] By the late 1980s, refinements like cycle sequencing—combining PCR amplification with termination—further streamlined workflows, minimizing template requirements and enabling direct sequencing of PCR products.[3] Japan's early investment in automation technologies from the 1980s positioned it as a leader in high-volume sequencing infrastructure.[41] The enhanced efficiency underpinned genome-scale initiatives in the 1990s. The Human Genome Project (HGP), planned since 1985 through international workshops, officially launched on October 1, 1990, under joint U.S. Department of Energy and National Institutes of Health oversight, targeting the 3.2 billion base pairs of human DNA via hierarchical shotgun sequencing with automated Sanger platforms.[42][43] Model organism projects followed: the Saccharomyces cerevisiae yeast genome, approximately 12 million base pairs, was sequenced by an international consortium and published in 1997 after completion in 1996, relying on automated fluorescent methods and yeast artificial chromosome mapping.[44] Escherichia coli's 4.6 million base pair genome was fully sequenced in 1997 using similar automated techniques.[44] Mid-1990s advancements included capillary electrophoresis systems, with Applied Biosystems introducing the ABI Prism 310 in 1995, replacing slab gels with narrower capillaries for faster runs (up to 600 bases) and higher resolution, processing one sample at a time but with reduced hands-on time.[37][36] Array-based capillaries later scaled to 96 or 384 lanes by the decade's end, supporting the HGP's goal of generating 1-2 million bases daily across centers.[40] These developments halved sequencing costs from about $1 per base in the early 1990s to $0.50 by 1998, enabling the era's focus on comprehensive genomic mapping over targeted gene analysis.[3]2000s Onward: High-Throughput Revolution and Cost Declines
The advent of next-generation sequencing (NGS) technologies in the mid-2000s marked a pivotal shift from labor-intensive Sanger sequencing to massively parallel approaches, enabling the simultaneous analysis of millions of DNA fragments and precipitating exponential declines in sequencing costs. The Human Genome Project, completed in 2003 at an estimated cost of approximately $2.7 billion using capillary-based Sanger methods, underscored the limitations of first-generation techniques for large-scale genomics, prompting investments like the National Human Genome Research Institute's (NHGRI) Revolutionary Sequencing Technologies program launched in 2004 to drive down costs by orders of magnitude.[45][46] Pioneering the NGS era, 454 Life Sciences introduced the Genome Sequencer GS 20 in 2005, employing pyrosequencing on emulsion PCR-amplified DNA beads captured in picoliter-scale wells, which generated up to 20 million bases per four-hour run—over 100 times the throughput of contemporary Sanger systems.[47] This platform demonstrated feasibility by sequencing the 580,000-base Mycoplasma genitalium genome in 2005, highlighting the potential for de novo assembly of microbial genomes without prior reference data.[47] Illumina followed in 2006 with the Genome Analyzer, utilizing sequencing-by-synthesis with reversible terminator chemistry on flow cells, initially yielding 1 gigabase per run and rapidly scaling to dominate the market due to its balance of throughput, accuracy, and cost-efficiency.[48] Applied Biosystems' SOLiD platform, commercialized around 2007, introduced ligation-based sequencing with di-base encoding for enhanced error detection, achieving high accuracy through two-base probe interrogation and supporting up to 60 gigabases per run in later iterations.[49] These innovations fueled a high-throughput revolution by leveraging clonal amplification (e.g., bridge or emulsion PCR) and array-based detection to process billions of short reads (typically 25-400 base pairs) in parallel, transforming genomics from a boutique endeavor to a data-intensive field. Applications expanded rapidly, including the 1000 Genomes Project launched in 2008 to catalog human genetic variation via NGS, which sequenced over 2,500 individuals and identified millions of variants.[50] Subsequent platforms like Illumina's HiSeq series (introduced 2010) further amplified output to terabases per run, while competition spurred iterative improvements in read length, error rates, and multiplexing.[48] By enabling routine whole-genome sequencing, NGS democratized access to genomic data, underpinning fields like population genetics, metagenomics, and personalized medicine, though challenges persisted in short-read alignment for repetitive regions and de novo assembly.[51] Sequencing costs plummeted as a direct consequence of these technological leaps and economies of scale, with NHGRI data showing the price per megabase dropping from about $5.60 in 2001 (adjusted for inflation) to under $0.05 by 2010, and per-genome costs falling from tens of millions to around $10,000 by 2010.[5] This Moore's Law-like trajectory, driven by increased parallelism, reagent optimizations, and market competition, reached approximately $1,000 per human genome by 2015 and continued declining to under $600 by 2023, far outpacing computational cost reductions and enabling projects like the UK Biobank's exome sequencing of 500,000 participants.[5][5] Despite these gains, comprehensive costs—including sample preparation, bioinformatics, and validation—remain higher than raw base-calling figures suggest, with ongoing refinements in library prep and error-correction algorithms sustaining the downward trend.[52]Classical Sequencing Methods
Maxam-Gilbert Chemical Cleavage
The Maxam–Gilbert method, introduced by Allan Maxam and Walter Gilbert in February 1977, represents the first practical technique for determining the nucleotide sequence of DNA through chemical cleavage at specific bases.[53] This approach cleaves terminally radiolabeled DNA fragments under conditions that partially modify purines or pyrimidines, generating a population of fragments terminating at each occurrence of the targeted base, which are then separated by size to reveal the sequence as a "ladder" on a gel.[33] Unlike enzymatic methods, it operates directly on double-stranded DNA without requiring prior strand separation for the cleavage reaction, though denaturation occurs during labeling and electrophoresis preparation.[21] The procedure begins with a purified DNA fragment of interest, typically 100–500 base pairs, produced via restriction enzyme digestion. One end of the double-stranded fragment is radiolabeled with phosphorus-32 using polynucleotide kinase and gamma-32P-ATP, followed by removal of the unlabeled strand via gel purification or exonuclease digestion to yield a single-stranded, end-labeled molecule.[21] Four parallel chemical reactions are then performed on aliquots of the labeled DNA, each designed to cleave preferentially at one or two bases:- G-specific cleavage: Dimethyl sulfate methylates the N7 position of guanine, rendering the phosphodiester backbone susceptible to hydrolysis by hot piperidine, which breaks the chain at ~1 in 20–50 guanines under controlled conditions.[53]
- A+G-specific cleavage: Formic acid depurinates adenine and guanine by protonating their glycosidic bonds, followed by piperidine-induced strand scission at apurinic sites.[21]
- T+C-specific cleavage: Hydrazine reacts with thymine and cytosine, forming hydrazones that piperidine cleaves, targeting pyrimidines.[53]
- C-specific cleavage: Hydrazine in the presence of 1–5 M sodium chloride selectively modifies cytosine, with piperidine completing the breaks, minimizing thymine interference.[21]
Sanger Chain-Termination Sequencing
The Sanger chain-termination method, also known as dideoxy sequencing, is an enzymatic technique for determining the nucleotide sequence of DNA. Developed by Frederick Sanger, Alan R. Coulson, and Simon Nicklen, it was first described in a 1977 paper published in the Proceedings of the National Academy of Sciences.[34] The method exploits the principle of DNA polymerase-mediated chain elongation, where synthesis terminates upon incorporation of a dideoxynucleotide triphosphate (ddNTP), a modified nucleotide lacking a 3'-hydroxyl group essential for phosphodiester bond formation.[3] This generates a population of DNA fragments of varying lengths, each ending at a specific nucleotide position corresponding to the incorporation of A-, C-, G-, or T-ddNTP.[54] In the original protocol, single-stranded DNA serves as the template, annealed to a radiolabeled oligonucleotide primer.[34] Four parallel reactions are performed, each containing DNA polymerase (typically from bacteriophage T7 or Klenow fragment of E. coli Pol I), all four deoxynucleotide triphosphates (dNTPs), and one of the four ddNTPs at a low concentration relative to dNTPs to ensure probabilistic termination.[21] Extension proceeds until a ddNTP is incorporated, producing fragments that are denatured and separated by size via polyacrylamide gel electrophoresis under denaturing conditions.[34] The resulting ladder of bands is visualized by autoradiography, with band positions revealing the sequence from the primer outward, typically reading up to 200-400 bases accurately.[3] Subsequent refinements replaced separate reactions with a single reaction using fluorescently labeled ddNTPs, each with a distinct dye for the four bases, enabling cycle sequencing akin to PCR for amplification and increased yield.[55] Fragments are then separated using capillary electrophoresis in automated sequencers, where laser excitation detects emission spectra to assign bases in real-time, extending read lengths to about 800-1000 base pairs with >99.9% accuracy per base.[21] This automation, commercialized in the late 1980s and 1990s, facilitated large-scale projects like the Human Genome Project, where Sanger sequencing provided finishing reads for gap closure despite the rise of parallel methods.[3] The method's fidelity stems from the high processivity and fidelity of DNA polymerase, minimizing errors beyond termination events, though limitations include bias toward GC-rich regions due to secondary structure and the need for cloning or PCR amplification of templates, which can introduce artifacts.[54] Despite displacement by high-throughput next-generation sequencing for bulk genomics, Sanger remains the gold standard for validating variants, sequencing short amplicons, and de novo assembly of small genomes owing to its precision and low error rate.[21]Next-Generation Sequencing (NGS)
Second-Generation: Amplification-Based Short-Read Methods
Second-generation DNA sequencing technologies, emerging in the mid-2000s, shifted from Sanger's serial chain-termination approach to massively parallel analysis of amplified DNA fragments, yielding short reads of 25 to 400 base pairs while drastically reducing costs per base sequenced.[56] These methods amplify template DNA via emulsion PCR (emPCR) or solid-phase bridge amplification to produce clonal clusters or bead-bound libraries, enabling simultaneous interrogation of millions of fragments through optical or electrical detection of nucleotide incorporation.[10] Amplification introduces biases, such as preferential enrichment of GC-balanced fragments, but facilitates signal amplification for high-throughput readout.[57] The Roche 454 platform, launched in 2005 as the first commercial second-generation system, employed pyrosequencing following emPCR amplification.[58] In this process, DNA libraries are fragmented, adapters ligated, and single molecules captured on beads within aqueous droplets in an oil emulsion for clonal amplification, yielding approximately 10^6 copies per bead.[59] Beads are then deposited into a fiber-optic slide with picoliter wells, where sequencing by synthesis occurs: unincorporated nucleotides trigger a luciferase-based reaction detecting pyrophosphate release as light flashes proportional to homopolymer length, with read lengths up to 400-1000 base pairs.[60] Despite higher error rates in homopolymers (up to 1.5%), 454 enabled rapid genome projects, such as the first individual human genome in 2008, but was discontinued in 2016 due to competition from cheaper alternatives.[58] Illumina's sequencing-by-synthesis (SBS), originating from Solexa technology acquired in 2007, dominates current short-read applications through bridge amplification on a flow cell.[48] DNA fragments with adapters hybridize to the flow cell surface, forming bridge structures that polymerase chain reaction (PCR) amplifies into dense clusters of ~1000 identical molecules each.[61] Reversible terminator nucleotides, each labeled with a distinct fluorophore, are flowed sequentially; incorporation is imaged, the terminator and label cleaved, allowing cyclic extension and base calling with per-base accuracy exceeding 99.9% for paired-end reads of 50-300 base pairs.[62] Systems like the HiSeq 2500 (introduced 2013) achieved terabase-scale output per run, fueling applications in whole-genome sequencing and transcriptomics, though PCR cycles can introduce duplication artifacts.[63] Applied Biosystems' SOLiD (Sequencing by Oligo Ligation and Detection), commercialized in 2007, uses emPCR-amplified bead libraries for ligation-based sequencing, emphasizing color-space encoding for error correction.[64] Adapter-ligated fragments are emulsified and amplified on magnetic beads, which are then deposited on a slide; di-base probes (two nucleotides long, with fluorophores indicating dinucleotide identity) are ligated iteratively, with degenerate positions enabling two-base resolution and query of each position twice across ligation cycles for >99.9% accuracy.[56] Reads averaged 50 base pairs, with two-base encoding reducing substitution errors but complicating analysis due to color-to-base translation.[64] The platform supported high-throughput variant detection but faded with Illumina's ascendancy. Ion Torrent, introduced by Life Technologies in 2010, integrates emPCR with semiconductor detection, bypassing optics for faster, cheaper runs.[65] Template DNA on Ion Sphere particles is amplified via emPCR, loaded onto a microwell array over ion-sensitive field-effect transistors; during SBS with unmodified nucleotides, proton release alters pH, generating voltage changes proportional to incorporated bases, yielding reads of 200-400 base pairs.[66] Lacking fluorescence, it avoids dye biases but struggles with homopolymers due to unquantified signal strength, with error rates around 1-2%.[67] Personal Genome Machine (PGM) models enabled benchtop sequencing of small genomes in hours.[68] These amplification-based methods collectively drove sequencing costs below $0.01 per megabase by 2015, enabling population-scale genomics, though short reads necessitate computational assembly and limit resolution of structural variants.[56]Third-Generation: Single-Molecule Long-Read Methods
Third-generation DNA sequencing encompasses single-molecule methods that sequence native DNA without amplification, allowing real-time detection of nucleotide incorporation or passage through a sensor, which yields reads typically exceeding 10 kilobases and up to megabases in length.[69] These approaches address limitations of second-generation short-read technologies, such as fragmentation-induced biases and challenges in resolving repetitive regions or structural variants.[70] Key platforms include Pacific Biosciences' Single Molecule Real-Time (SMRT) sequencing and Oxford Nanopore Technologies' (ONT) nanopore sequencing, both commercialized in the early 2010s.[71] SMRT sequencing, developed by Pacific Biosciences (founded in 2004), employs zero-mode waveguides—nanoscale wells that confine observation volumes to enable real-time fluorescence detection of DNA polymerase activity on surface-immobilized templates.[72] The process uses a double-stranded DNA template ligated into a hairpin-loop structure called a SMRTbell, where a phi29 DNA polymerase incorporates fluorescently labeled reversible-terminator nucleotides, with each base's distinct emission spectrum captured via pulsed laser excitation.[71] Initial raw read accuracies were around 85-90% due to polymerase processivity limits and signal noise, but circular consensus sequencing (CCS), introduced later, generates high-fidelity (HiFi) reads exceeding 99.9% accuracy by averaging multiple passes over the same molecule, with read lengths up to 20-30 kilobases.[73] The first commercial instrument, the PacBio RS, launched in 2010, followed by the Sequel system in 2015 and Revio in 2022, which increased throughput to over 1 terabase per run via higher ZMW density.[71] ONT sequencing passes single-stranded DNA or RNA through a protein nanopore (typically engineered variants of Mycobacterium smegmatis porin A) embedded in a membrane, controlled by a helicase or polymerase motor protein, while measuring disruptions in transmembrane ionic current as bases transit the pore's vestibule.[70] Each nucleotide or dinucleotide motif produces a unique current signature, decoded by basecalling algorithms; this label-free method also detects epigenetic modifications like 5-methylcytosine directly from native strands.[74] Development began in the mid-2000s, with the portable MinION device released for early access in 2014, yielding initial reads up to 100 kilobases, though raw error rates hovered at 5-15% from homopolymer inaccuracies and signal drift.[75] Subsequent flow cells like PromethION, deployed since 2018, support ultra-long reads exceeding 2 megabases and outputs up to 290 gigabases per run, with adaptive sampling and improved chemistry reducing errors to under 5% in Q20+ modes by 2023.[76] These methods excel in de novo genome assembly of complex, repeat-rich organisms—such as the human genome's challenging centromeric regions—and haplotype phasing, where long reads span variants separated by hundreds of kilobases, outperforming short-read approaches that often require hybrid assemblies.[77] They also enable direct RNA sequencing for isoform resolution and variant detection in transcripts up to full-length.[70] However, raw per-base error rates remain higher than second-generation platforms (though mitigated by consensus), and early instruments suffered from lower throughput and higher costs per gigabase, limiting scalability for population-scale projects until recent hardware advances.[78] Despite these, third-generation technologies have driven breakthroughs in metagenomics and structural variant calling, with error-corrected assemblies achieving near-complete bacterial genomes and improved eukaryotic contiguity.[79]Emerging and Specialized Sequencing Techniques
Nanopore and Tunneling-Based Approaches
Nanopore sequencing detects the sequence of DNA or RNA by measuring disruptions in an ionic current as single molecules translocate through a nanoscale pore embedded in a membrane.[80] The pore, typically a protein such as α-hemolysin or engineered variants like Mycobacterium smegmatis porin, or solid-state alternatives, allows ions to flow while the nucleic acid strand passes through, with each base causing a characteristic blockade in current amplitude and duration.[75] This approach enables real-time, label-free sequencing without amplification, producing reads often exceeding 100,000 bases in length, which facilitates resolving repetitive regions and structural variants intractable to short-read methods.[70] Oxford Nanopore Technologies has commercialized this method since 2014 with devices like the portable MinION and high-throughput PromethION, achieving throughputs up to 290 Gb per flow cell as of 2023.[80] Early implementations suffered from raw read accuracies of 85-92%, limited by noisy signals and basecalling errors, particularly in homopolymers.[81] Iterative improvements, including dual-reader pores in R10 flow cells and advanced algorithms like Dorado basecallers, have elevated single-read accuracy to over 99% for DNA by 2024, with Q20+ consensus modes yielding near-perfect assemblies when combining multiple reads.[82] These advancements stem from enhanced motor proteins for controlled translocation at ~450 bases per second and machine learning for signal interpretation, reducing systematic errors in RNA sequencing to under 5% in optimized protocols.[83] Tunneling-based approaches leverage quantum mechanical electron tunneling to identify bases by their distinct transverse conductance signatures as DNA threads through a nanogap or junction, offering potentially higher resolution than ionic current alone.[84] In configurations like gold nanogaps or graphene edge junctions, electrons tunnel across electrodes separated by 1-2 nm, with current modulation varying by base-specific electronic orbitals—A-T pairs exhibit higher tunneling probabilities than G-C due to differing HOMO-LUMO gaps.[85] Research prototypes integrate this with nanopores, using self-aligned transverse junctions to correlate tunneling signals with translocation events, achieving >93% detection yield in DNA passage experiments as of 2021.[86] Developments in tunneling detection include machine learning-aided quantum transport models, which classify artificial DNA sequences with unique current fingerprints, as demonstrated in 2025 simulations predicting base discrimination at zeptojoule sensitivities.[87] Combined quantum tunneling and dielectrophoretic trapping in capillary nanoelectrodes enable standalone probing without conductive substrates, though signal-to-noise challenges persist in wet environments.[88] Unlike mature nanopore systems, tunneling methods remain largely experimental, with no widespread commercial platforms by 2025, due to fabrication precision demands and integration hurdles, but hold promise for ultra-fast, amplification-free sequencing if scalability improves.[89]Sequencing by Hybridization and Mass Spectrometry
Sequencing by hybridization (SBH) determines DNA sequences by hybridizing fragmented target DNA to an array of immobilized oligonucleotide probes representing all possible short sequences (n-mers, typically 8-10 bases long), identifying binding patterns to reconstruct the original sequence computationally.[90] Proposed in the early 1990s, SBH leverages the specificity of Watson-Crick base pairing under controlled stringency conditions to detect complementary subsequences, with positive hybridization signals indicating presence in the target.[91] Early demonstrations achieved accurate reconstruction of up to 100 base pairs using octamer and nonamer probes in independent reactions, highlighting its potential for parallel analysis without enzymatic extension.[92] Key advancements include positional SBH (PSBH), introduced in 1994, which employs duplex probes with single-base mismatches to resolve ambiguities and extend readable lengths by encoding positional information directly in hybridization spectra.[93] Microchip-based implementations by 1996 enabled efficient scaling with oligonucleotide arrays, increasing probe density and throughput for de novo sequencing or resequencing known regions.[94] Ligation-enhanced variants, developed around 2002, combine short probes into longer ones via enzymatic joining, reducing the exponential probe set size (e.g., from millions for 10-mers to tens for extended reads) while improving specificity for complex samples up to thousands of bases.[95] Despite these, SBH's practical utility remains limited to short fragments or validation due to challenges like cross-hybridization errors from near-perfect matches, incomplete coverage in repetitive regions, and the combinatorial explosion of probes required for long sequences, necessitating robust algorithms for spectrum reconstruction.[96] Applications include high-throughput genotyping and fingerprinting of mixed DNA/RNA samples, though it has been largely supplanted by amplification-based methods for genome-scale work.[97] Mass spectrometry (MS)-based DNA sequencing measures the mass-to-charge ratio of ionized DNA fragments to infer sequence, often adapting Sanger dideoxy termination by replacing electrophoretic separation with MS detection via techniques like matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) or electrospray ionization (ESI).[98] Pioneered in the mid-1990s, this approach generates termination products in a single tube using biotinylated dideoxynucleotides for purification, then analyzes fragment masses to deduce base order, offering advantages over gel-based methods such as elimination of dye-induced mobility shifts and faster readout (seconds per sample versus hours).[99] By 1998, MALDI-TOF protocols enabled reliable sequencing of up to 50-100 bases with fidelity comparable to traditional Sanger, particularly for oligonucleotides, through delayed extraction modes to enhance resolution.[100] Applications focus on short-read validation, SNP genotyping, and mutation detection rather than de novo assembly, as MS excels in precise mass differentiation for small variants (e.g., single-base substitutions via ~300 Da shifts) but struggles with longer fragments due to resolution limits (typically <200 bases) and adduct formation from salts or impurities requiring extensive sample cleanup.[101] Challenges include low ionization efficiency for large polyanionic DNA, spectral overlap in heterogeneous mixtures, and sensitivity to sequence-dependent fragmentation, restricting throughput compared to optical methods; tandem MS (MS/MS) extensions for double-stranded DNA have been explored but remain niche.[102] Despite potential for automation in diagnostics, MS sequencing has not scaled to high-throughput genomes, overshadowed by NGS since the 2000s, though it persists for confirmatory assays in forensics and clinical validation.[103]Recent Innovations: In Situ and Biochemical Encoding Methods
In situ genome sequencing (IGS) enables direct readout of genomic DNA sequences within intact cells or tissues, preserving spatial context without extraction. Introduced in 2020, IGS constructs sequencing libraries in situ via transposition and amplification, followed by barcode hybridization and optical decoding to resolve DNA sequences and chromosomal positions at subcellular resolution.[104] This method has mapped structural variants and copy number alterations in cancer cell lines, achieving ~1 kb resolution for loci-specific sequencing.[105] A 2025 advancement, expansion in situ genome sequencing (ExIGS), integrates IGS with expansion microscopy to enhance spatial resolution beyond the diffraction limit. By embedding samples in a swellable hydrogel that expands isotropically by ~4.5-fold, ExIGS localizes sequenced DNA loci and nuclear proteins at ~60 nm precision, enabling quantification of 3D genome organization disruptions.[106] Applied to progeria models, ExIGS revealed lamin A/C mutations cause locus-specific radial repositioning and altered chromatin interactions, with affected loci shifting ~500 nm outward from the nuclear center compared to wild-type cells.[107] This technique supports multimodal imaging, combining DNA sequence data with protein immunofluorescence to link genomic aberrations to nuclear architecture defects.[108] Biochemical encoding methods innovate by transforming native DNA sequences into amplified, decodable polymers prior to readout. Roche's Sequencing by Expansion (SBX), unveiled in February 2025, employs enzymatic synthesis to encode target DNA into Xpandomers—cross-linked, expandable polymers that replicate the original sequence at high fidelity.[109] This approach mitigates amplification biases in traditional NGS by generating uniform, high-density signals for short-read sequencing, potentially reducing error rates in low-input samples to below 0.1%.[110] SBX's biochemical cascade involves template-directed polymerization and reversible termination, enabling parallel processing of millions of fragments with claimed 10-fold cost efficiency over prior amplification schemes.[111] Proximity-activated DNA scanning encoded sequencing (PADSES), reported in April 2025, uses biochemical tags to encode spatial proximity data during interaction mapping. This method ligates barcoded adapters to interacting DNA loci in fixed cells, followed by pooled sequencing to resolve contact frequencies at single-molecule scale, achieving >95% specificity for enhancer-promoter pairs in human cell lines.[112] Such encoding strategies extend beyond linear sequencing to capture higher-order genomic interactions, informing causal regulatory mechanisms with empirical resolution of <10 kb.[113]Data Processing and Computational Frameworks
Sequence Assembly: Shotgun and De Novo Strategies
Sequence assembly reconstructs the continuous DNA sequence from short, overlapping reads generated during shotgun sequencing, a process essential for de novo genome reconstruction without a reference. In shotgun sequencing, genomic DNA is randomly fragmented into small pieces, typically 100–500 base pairs for next-generation methods or longer for Sanger-era approaches, and each fragment is sequenced to produce reads with sufficient overlap for computational reassembly. This strategy enables parallel processing of millions of fragments, scaling to large genomes, but requires high coverage—often 10–30× the genome size—to ensure overlaps span the entire sequence.[114][115] The whole-genome shotgun (WGS) approach, a hallmark of modern assembly, omits prior physical mapping by directly sequencing random fragments and aligning them via overlaps, contrasting with hierarchical methods that first construct and map clone libraries like bacterial artificial chromosomes (BACs). WGS was pivotal in the Celera Genomics effort, which produced a draft human genome in 2001 using approximately 5× coverage from Sanger reads, demonstrating feasibility for complex eukaryotic genomes despite initial skepticism over repeat resolution. Advantages include reduced labor and cost compared to mapping-based strategies, though limitations arise in repetitive regions exceeding read lengths, where ambiguities lead to fragmented contigs. Mate-pair or paired-end reads, linking distant fragments, aid scaffolding by providing long-range information to order contigs into scaffolds.[116][114][117] De novo assembly algorithms process shotgun reads without alignment to a reference, employing two primary paradigms: overlap-layout-consensus (OLC) and de Bruijn graphs (DBG). OLC detects pairwise overlaps between reads (e.g., via suffix trees or minimizers), constructs an overlap graph with reads as nodes and overlaps as edges, lays out paths representing contigs, and derives consensus sequences by multiple sequence alignment; it excels with longer, lower-coverage reads as in third-generation sequencing, but computational intensity scales poorly with short-read volume. DBG, optimized for short next-generation reads, decomposes reads into k-mers (substrings of length k), builds a directed graph where nodes represent (k-1)-mers and edges denote k-mers, then traverses via an Eulerian path to reconstruct the sequence, inherently handling errors through coverage-based tipping. DBG mitigates sequencing noise better than OLC for high-throughput data but struggles with uneven coverage or low-complexity repeats forming tangled subgraphs. Hybrid approaches combine both for improved contiguity, as seen in assemblers like Canu for long reads.[118][119][120] Challenges in both strategies include resolving structural variants, heterozygosity in diploids causing haplotype bubbles, and chimeric assemblies from contaminants; metrics like N50 contig length (where 50% of the genome lies in contigs of that length or longer) and BUSCO completeness assess quality, with recent long-read advances pushing N50s beyond megabases for human genomes. Empirical data from benchmarks show DBG outperforming OLC in short-read accuracy for bacterial genomes (e.g., >99% identity at 100× coverage), while OLC yields longer scaffolds in eukaryotic projects like the 2000 Drosophila assembly. Ongoing innovations, such as compressed data structures, address scalability for terabase-scale datasets.[118][121][114]Quality Control: Read Trimming and Error Correction
Quality control in next-generation sequencing (NGS) pipelines begins with read trimming to excise artifacts and low-confidence bases, followed by error correction to mitigate systematic and random sequencing inaccuracies, thereby enhancing data reliability for assembly and variant detection. Raw reads from platforms like Illumina often exhibit declining base quality toward the 3' ends due to dephasing and incomplete extension cycles, with Phred scores (Q-scores) quantifying error probabilities as Q = -10 log10(P), where P is the mismatch probability. Trimming preserves usable high-quality portions while discarding noise, typically reducing false positives in downstream analyses by 10-20% in alignment-based tasks.[122] Adapter trimming targets synthetic sequences ligated during library preparation, which contaminate reads when fragments are shorter than read lengths; tools like Cutadapt or Trimmomatic scan for exact or partial matches using seed-and-extend algorithms, removing them to prevent misalignment artifacts. Quality-based trimming employs heuristics such as leading/trailing clip thresholds (e.g., Q < 3) and sliding window filters (e.g., 4-base window with average Q < 20), as implemented in Trimmomatic, which processes paired-end data while enforcing minimum length cutoffs (e.g., 36 bases) to retain informative reads. Evaluations across datasets show these methods boost mappable read fractions from 70-85% in untrimmed data to over 95%, though aggressive trimming risks over-removal in low-diversity libraries.[122][123] Error correction algorithms leverage data redundancy from high coverage (often >30x) to resolve substitutions, insertions, and deletions arising from polymerase infidelity, optical noise, or phasing errors, which occur at rates of 0.1-1% per base in short-read NGS. Spectrum-based methods, such as those in Quake or BFC, construct k-mer frequency histograms to identify erroneous rare k-mers and replace them with high-frequency alternatives, achieving up to 70% error reduction in high-coverage microbial genomes. Overlap-based correctors like Muskrat or CARE align short windows between reads using suffix arrays or Burrows-Wheeler transforms to derive consensus votes, excelling in detecting clustered errors but scaling poorly with dataset size (O(n^2) time complexity). Hybrid approaches, integrating short-read consensus with long-read scaffolding, have demonstrated superior indel correction (error rates dropping from 1-5% to <0.5%) in benchmarks using UMI-tagged high-fidelity data.[124][125] Recent advancements emphasize context-aware correction, such as CARE's use of read neighborhoods for haplotype-informed fixes, reducing chimeric read propagation in variant calling pipelines. Benchmarks indicate that no single algorithm universally outperforms others across error profiles—e.g., k-mer methods falter in repetitive regions (>1% error persistence)—necessitating tool selection based on read length, coverage, and genome complexity, with post-correction Q-score recalibration via tools like GATK's PrintReads further refining outputs. Over-correction risks inflating coverage biases, so validation against gold-standard datasets remains essential, as uncorrected errors propagate to inflate variant false discovery rates by 2-5-fold in low-coverage scenarios.[126][124][127]Bioinformatics Pipelines and Scalability Challenges
Bioinformatics pipelines in next-generation sequencing (NGS) encompass automated, modular workflows designed to process vast quantities of raw sequence data into actionable insights, such as aligned reads, variant calls, and functional annotations. These pipelines typically begin with quality assessment using tools like FastQC to evaluate read quality metrics, followed by adapter trimming and filtering with software such as Trimmomatic or Cutadapt to remove low-quality bases and artifacts. Subsequent alignment of reads to a reference genome employs algorithms like BWA-MEM or Bowtie2, generating sorted BAM files that capture mapping positions and discrepancies. Variant calling then utilizes frameworks such as GATK or DeepVariant to identify single nucleotide variants, insertions/deletions, and structural alterations based on coverage depth and allele frequencies.[10][128] Further pipeline stages include post-processing for duplicate removal, base quality score recalibration, and annotation against databases like dbSNP or ClinVar to classify variants by pathogenicity and population frequency. For specialized analyses, such as RNA-seq or metagenomics, additional modules integrate tools like STAR for splice-aware alignment or QIIME2 for taxonomic profiling. Workflow management systems, including Nextflow, Snakemake, or Galaxy, orchestrate these steps, ensuring reproducibility through containerization with Docker or Singularity and declarative scripting in languages like WDL or CWL. In clinical settings, pipelines must adhere to validation standards from organizations like AMP and CAP, incorporating orthogonal confirmation for high-impact variants to mitigate false positives from algorithmic biases.[10][128] Scalability challenges emerge from the sheer volume and complexity of NGS data, where a single human whole-genome sequencing (WGS) sample at 30× coverage generates approximately 150 GB of aligned data, escalating to petabytes for population-scale studies. Computational demands intensify during alignment and variant calling, which can require hundreds of CPU cores and terabytes of RAM; for instance, GATK processing of an 86 GB dataset completes in under 3 hours on 512 cores, but bottlenecks persist in I/O operations and memory-intensive joint genotyping across cohorts. Storage and transfer costs compound issues, with raw FASTQ files alone demanding petabyte-scale infrastructure, prompting reliance on high-performance computing (HPC) clusters, cloud platforms like AWS, or distributed frameworks such as Apache Spark for elastic scaling.[129][130] To address these, pipelines leverage parallelization strategies like MapReduce for data partitioning and GPU acceleration for read alignment, reducing processing times by factors of 10-50× in benchmarks of WGS datasets. However, challenges in reproducibility arise from version dependencies and non-deterministic parallel execution, necessitating provenance tracking and standardized benchmarks. Emerging solutions include federated learning for privacy-preserving analysis and optimized formats like CRAM for compressed storage, yet the trade-offs between accuracy, speed, and cost remain critical, particularly for resource-limited labs handling increasing throughput from platforms generating billions of reads per run.[130][131]Applications
Fundamental Research: Molecular and Evolutionary Biology
DNA sequencing has enabled precise annotation of genomes, distinguishing protein-coding genes from regulatory elements such as promoters, enhancers, and non-coding RNAs, which comprise over 98% of the human genome.[2] This capability underpins molecular studies of gene regulation, where techniques like chromatin immunoprecipitation followed by sequencing (ChIP-seq) map transcription factor binding sites and histone modifications to reveal epigenetic controls on expression.[10] For example, sequencing of model organisms like Saccharomyces cerevisiae in 1996 identified approximately 6,000 genes, facilitating functional genomics experiments that linked sequence variants to phenotypic traits such as metabolic pathways.[132] In protein-DNA interactions and pathway elucidation, sequencing supports CRISPR-Cas9 off-target analysis, quantifying unintended edits through targeted amplicon sequencing to assess editing specificity, which reached error rates below 0.1% in optimized protocols by 2018.[133] RNA sequencing (RNA-seq) extends this to transcriptomics, quantifying alternative splicing and isoform diversity; a 2014 study of human cell lines revealed over 90% of multi-exon genes undergo splicing, challenging prior estimates and informing models of post-transcriptional regulation.[134] Single-molecule sequencing further dissects molecular heterogeneity, as in long-read approaches resolving full-length transcripts without assembly artifacts, enhancing understanding of RNA secondary structures and pseudogene interference.[135] For evolutionary biology, DNA sequencing drives phylogenomics by generating alignments of orthologous genes across taxa, enabling maximum-likelihood tree inference that resolves divergences like the animal kingdom's basal branches with bootstrap support exceeding 95% in datasets of over 1,000 genes.[136] Comparative genomics identifies conserved synteny blocks, such as those spanning 40% of human-mouse genomes despite 75 million years of divergence, indicating purifying selection on regulatory architectures.[137] Sequencing ancient DNA, including Neanderthal genomes from 2010 yielding 1.3-fold coverage, quantifies admixture events contributing 1-4% archaic ancestry in non-African populations, while site-frequency spectrum analysis detects positive selection signatures, like in MHC loci under pathogen-driven evolution.[132] These methods refute gradualist models by revealing punctuated gene family expansions, such as transposon proliferations accounting for 45% of mammalian genome size variation.[138]Clinical and Diagnostic Uses: Precision Medicine and Oncology
DNA sequencing technologies, especially next-generation sequencing (NGS), underpin precision medicine by enabling the identification of individual genetic variants that inform tailored therapeutic strategies, reducing reliance on empirical treatment approaches. In oncology, NGS facilitates comprehensive tumor genomic profiling, detecting somatic mutations, gene fusions, and copy number alterations that serve as biomarkers for targeted therapies, immunotherapy response, or clinical trial eligibility. For instance, FDA-approved NGS-based companion diagnostics, such as those for EGFR, ALK, and BRAF alterations, guide the selection of inhibitors like osimertinib or dabrafenib-trametinib in non-small cell lung cancer and melanoma, respectively, improving progression-free survival rates compared to standard chemotherapy.[139][140][141] Clinical applications extend to solid and hematologic malignancies, where whole-exome or targeted gene panel sequencing analyzes tumor DNA to uncover actionable drivers, with studies reporting that 30-40% of advanced cancer patients harbor variants matchable to approved therapies.[142] In 2024, whole-genome sequencing of solid tumors demonstrated high sensitivity for detecting low-frequency mutations and structural variants, correlating with treatment responsiveness in real-world cohorts.[143] Liquid biopsy techniques, involving cell-free DNA sequencing from blood, enable non-invasive monitoring of tumor evolution, minimal residual disease detection post-treatment, and early identification of resistance mechanisms, such as MET amplifications emerging during EGFR inhibitor therapy.[144] The integration of NGS into oncology workflows has accelerated since FDA authorizations of mid-sized panels in 2017-2018, expanding to broader comprehensive genomic profiling tests by 2025, which analyze hundreds of genes across tumor types agnostic to histology.[139][145] Retrospective analyses confirm that NGS-informed therapies yield superior outcomes in gastrointestinal cancers, with matched treatments extending median overall survival by months in biomarker-positive subsets.[146] These diagnostic uses also support pharmacogenomics, predicting adverse reactions to chemotherapies like irinotecan based on UGT1A1 variants, thereby optimizing dosing and minimizing toxicity.[147] Despite variability in panel coverage and interpretation, empirical data from large cohorts underscore NGS's causal role in shifting oncology from one-size-fits-all paradigms to genotype-driven interventions.[148]Forensic, Ancestry, and Population Genetics
Next-generation sequencing (NGS) technologies have transformed forensic DNA analysis by enabling the parallel interrogation of multiple markers, including short tandem repeats (STRs), single nucleotide polymorphisms (SNPs), and mitochondrial DNA variants, from challenging samples such as degraded or trace evidence.[149] Unlike capillary electrophoresis methods limited to 20-24 STR loci in systems like CODIS, NGS supports massively parallel amplification and sequencing, improving resolution for mixture deconvolution—where multiple contributors are present—and kinship determinations in cases lacking direct reference samples.[150] Commercial panels, such as the ForenSeq system, integrate over 200 markers for identity, lineage, and ancestry inference, with validation studies demonstrating error rates below 1% for allele calls in controlled conditions.[150] These advances have facilitated identifications in cold cases, such as the 2021 resolution of the Golden State Killer through investigative genetic genealogy, though NGS adoption remains constrained by validation standards and computational demands for variant calling.[150] In ancestry testing, DNA sequencing underpins advanced biogeographical estimation by analyzing genome-wide variants against reference panels of known ethnic origins, though most direct-to-consumer services rely on targeted SNP genotyping arrays scanning ~700,000 sites rather than complete sequencing of the 3 billion base pairs.[151] Whole-genome sequencing (WGS), when applied, yields higher granularity for admixture mapping—quantifying proportions of continental ancestry via linkage disequilibrium blocks—and relative matching to ancient DNA, as in studies aligning modern sequences to Neolithic samples for tracing migrations over millennia.[152] Inference accuracy varies, with European-descent references yielding median errors of 5-10% for continental assignments, but underrepresentation in non-European databases leads to inflated uncertainty for African or Indigenous American ancestries, as evidenced by cross-validation against self-reported pedigrees.[152] Services offering WGS, such as those sequencing 100% of the genome, enhance detection of rare variants for distant relatedness but require imputation for unsequenced regions and face challenges from recombination breaking long-range haplotypes.[153] Population genetics leverages high-throughput sequencing to assay allele frequencies across cohorts, enabling inferences of demographic events like bottlenecks or expansions through site frequency spectrum analysis and coalescent modeling.[154] For instance, reduced representation sequencing of pooled samples from wild populations captures thousands of SNPs per individual at costs under $50 per genome, facilitating studies of local adaptation via scans for selective sweeps in genes like those for lactase persistence.[155] In humans, large-scale efforts have sequenced over 100,000 exomes to map rare variant burdens differing by ancestry, revealing causal alleles for traits under drift or selection, while ancient DNA integration via sequencing of ~5,000 prehistoric genomes has quantified Neanderthal admixture at 1-2% in non-Africans.[155] These methods demand robust error correction for low-frequency variants, with pipelines like GATK achieving >99% call accuracy, but sampling biases toward urban or admixed groups can skew inferences of neutral diversity metrics such as π (nucleotide diversity) by up to 20% in underrepresented populations.[156]Environmental and Metagenomic Sequencing
Environmental and metagenomic sequencing refers to the direct extraction and analysis of genetic material from environmental samples, such as soil, water, sediment, or air, to characterize microbial and multicellular communities without isolating individual organisms. This approach, termed metagenomics, was first conceptualized in the mid-1980s by Norman Pace, who advocated sequencing ribosomal RNA genes from uncultured microbes to assess diversity.[157] The field advanced with the 1998 coining of "metagenome" by Handelsman and colleagues, describing the collective genomes in a habitat.[158] A landmark 2004 study by Craig Venter's team sequenced Sargasso Sea microbial DNA using Sanger methods, identifying over 1,800 new species and 1.2 million novel genes, demonstrating the vast unculturable microbial diversity.[157] Two primary strategies dominate: targeted amplicon sequencing, often of the 16S rRNA gene for prokaryotes, which profiles taxonomic composition but misses functional genes and underrepresents rare taxa due to PCR biases and primer mismatches; and shotgun metagenomics, which randomly fragments and sequences total DNA for both taxonomy and metabolic potential, though it demands higher throughput and computational resources.[159][160] Shotgun approaches, enabled by next-generation sequencing since the 2000s, yield deeper insights—identifying more taxa and enabling gene annotation—but generate vast datasets challenging assembly due to strain-level variation and uneven coverage.[161] Environmental DNA (eDNA) sequencing extends this to macroorganisms, detecting shed genetic traces for non-invasive biodiversity surveys, as in aquatic systems where fish or amphibians leave DNA in water persisting hours to days.[162] Applications span ecosystem monitoring and discovery: metagenomics has mapped ocean microbiomes, as in the 2007-2013 Tara Oceans expedition, which cataloged 35,000 operational taxonomic units and millions of genes influencing carbon cycling.[157] In terrestrial environments, soil metagenomes reveal nutrient-cycling microbes, aiding agriculture by identifying nitrogen-fixing bacteria.[163] eDNA enables rapid invasive species detection, such as Asian carp in U.S. rivers via mitochondrial markers, outperforming traditional netting in sensitivity.[164] Functionally, it uncovers enzymes for bioremediation, like plastic-degrading plastics from marine samples, and antibiotics from uncultured bacteria, addressing antimicrobial resistance.[165] Challenges persist: extraction biases favor certain taxa (e.g., Gram-positive bacteria underrepresented in soil), contamination from reagents introduces false positives, and short reads hinder resolving complex assemblies in high-diversity samples exceeding 10^6 species per gram of soil.[166] Incomplete reference genomes limit annotation, with only ~1% of microbial species cultured, inflating unknown sequences to 50-90% in many datasets.[167] Computational pipelines require binning tools like MetaBAT for metagenome-assembled genomes, but scalability lags for terabase-scale projects, necessitating hybrid long-read approaches for better contiguity.[168] Despite these, metagenomics has transformed ecology by quantifying causal microbial roles in processes like methane production, grounded in empirical sequence-function links rather than culture-dependent assumptions.[163]Agricultural and Industrial Biotechnology
DNA sequencing technologies have revolutionized agricultural biotechnology by enabling the precise identification of genetic markers linked to traits such as yield, disease resistance, and environmental tolerance in crops and livestock. In plant breeding, marker-assisted selection (MAS) leverages DNA sequence data to select progeny carrying specific alleles without relying solely on phenotypic evaluation, reducing breeding cycles from years to months in some cases. For instance, sequencing of crop genomes like maize and rice has revealed quantitative trait loci (QTLs) controlling kernel size and drought tolerance, allowing breeders to introgress favorable variants into elite lines.[169][170][171] In livestock applications, whole-genome sequencing supports genomic selection, where dense SNP markers derived from sequencing predict breeding values for traits like milk production in cattle or growth rates in poultry, achieving accuracy rates up to 80% higher than traditional methods. This approach has been implemented in Brazil's cattle industry since the mid-2010s, enhancing herd productivity through targeted matings informed by sequence variants. Similarly, in crop wild relatives, transcriptome sequencing identifies novel alleles for traits absent in domesticated varieties, aiding introgression for climate-resilient hybrids, as demonstrated in efforts to bolster disease resistance in wheat.[172][173] In industrial biotechnology, DNA sequencing underpins metabolic engineering of microorganisms for enzyme production and biofuel synthesis by mapping pathways and optimizing gene clusters. For biofuel applications, sequencing of lignocellulolytic bacteria, such as those isolated from extreme environments, has identified thermostable cellulases with activity optima above 70°C, improving saccharification efficiency in ethanol production by up to 50% compared to mesophilic counterparts. Sequencing also facilitates directed evolution of strains, as seen in yeast engineered for isobutanol yields exceeding 100 g/L through iterative variant analysis.[174][175][176] These advancements rely on high-throughput sequencing to generate variant maps, though challenges persist in polyploid crops where assembly errors can confound allele calling, necessitating hybrid long-read approaches for accurate haplotype resolution. Overall, sequencing-driven strategies have increased global crop yields by an estimated 10-20% in sequenced staples since 2010, while industrial processes benefit from reduced development timelines for scalable biocatalysts.[177][178]Technical Limitations and Engineering Challenges
Accuracy, Coverage, and Read Length Constraints
Accuracy in DNA sequencing refers to the per-base error rate, which varies significantly across technologies and directly impacts variant calling reliability. Short-read platforms like Illumina achieve raw per-base accuracies of approximately 99.9%, corresponding to error rates of 0.1–1% before correction, with errors primarily arising from base-calling algorithms and PCR amplification artifacts.[179] Long-read technologies, such as Pacific Biosciences (PacBio) and Oxford Nanopore, historically exhibited higher error rates—up to 10–15% for early iterations—due to challenges in signal detection from single-molecule templates, though recent advancements have reduced these to under 1% with consensus polishing.[10] These errors are mitigated through increased coverage depth, where consensus from multiple overlapping reads enhances overall accuracy; for instance, error rates for non-reference genotype calls drop to 0.1–0.6% at sufficient depths.[180] Coverage constraints involve both depth (average number of reads per genomic position) and uniformity, essential for detecting low-frequency variants and avoiding false negatives. For human whole-genome sequencing, 30–50× average depth is standard to achieve >99% callable bases with high confidence, as lower depths increase uncertainty in heterozygous variant detection.[181] De novo assembly demands higher depths of 50–100× to resolve ambiguities in repetitive regions.[182] Uniformity is compromised by biases, notably GC content bias, where extreme GC-rich or AT-rich regions receive 20–50% fewer reads due to inefficient amplification and sequencing chemistry, leading to coverage gaps that can exceed 10% of the genome in biased samples.[183][184] Read length constraints impose trade-offs between resolution of complex genomic structures and per-base fidelity. Short reads (typically 100–300 base pairs) excel in high-throughput applications but fail to span repetitive elements longer than their length, complicating assembly and structural variant detection, where up to 34% of disease-associated variants involve large insertions or duplications missed by short-read data.[185] Long reads (>10,000 base pairs) overcome these by traversing repeats and resolving haplotypes, enabling superior de novo assembly, yet their lower raw accuracy necessitates hybrid approaches combining long reads for scaffolding with short reads for polishing.[186][187] These limitations persist despite engineering improvements, as fundamental biophysical constraints in polymer translocation and base detection limit ultra-long read fidelity without consensus strategies.[188]Sample Preparation Biases and Contamination Risks
Sample preparation for next-generation sequencing (NGS) involves DNA extraction, fragmentation, end repair, adapter ligation, and often PCR amplification to generate sequencing libraries. These steps introduce systematic biases that distort representation of the original genomic material. GC content bias, a prominent issue, manifests as uneven read coverage correlating with regional GC percentage, typically underrepresenting high-GC (>60%) and extremely low-GC (<30%) regions due to inefficient polymerase extension and denaturation during PCR.[183] This bias arises primarily from enzymatic inefficiencies in library preparation kits, with studies demonstrating up to 10-fold coverage variation across GC extremes in human genome sequencing.[189] PCR-free protocols reduce but do not eliminate this effect, as fragmentation methods like sonication or tagmentation (e.g., Nextera) exhibit platform-specific preferences, with Nextera showing pronounced undercoverage in low-GC areas.[190][191] Additional biases stem from priming strategies and fragment size selection. Random hexamer priming during reverse transcription or library amplification favors certain motifs, leading to overrepresentation of AT-rich starts in reads.[192] Size selection via gel electrophoresis or bead-based purification skews toward preferred fragment lengths (often 200-500 bp), underrepresenting repetitive or structurally complex regions like centromeres. In metagenomic applications, these biases exacerbate under-detection of low-abundance taxa with atypical GC profiles, with library preparation alone accounting for up to 20% deviation in community composition estimates.[193] Mitigation strategies include bias-correction algorithms post-sequencing, such as lowess normalization, though they cannot recover lost signal from underrepresented regions.[194] Contamination risks during sample preparation compromise data integrity, particularly for low-input or ancient DNA samples where exogenous sequences can dominate. Commercial DNA extraction kits and reagents frequently harbor microbial contaminants, with one analysis detecting bacterial DNA from multiple phyla in over 90% of tested kits, originating from manufacturing environments and persisting through ultra-clean processing.[195] Pre-amplification steps amplify these contaminants exponentially, introducing chimeric sequences that mimic true variants in downstream analyses.[196] In multiplexed Illumina sequencing, index hopping—caused by free adapter dimers ligating during bridge amplification—results in 0.1-1% of reads misassigned to incorrect samples, with rates reaching 3% under high cluster density or incomplete library cleanup.[197][198] Cross-sample contamination from pipetting aerosols or shared workspaces further elevates risks, potentially yielding false positives in rare variant detection at frequencies as low as 0.01%.[199] Dual unique indexing and dedicated cleanroom protocols minimize these issues, though empirical validation via spike-in controls remains essential for quantifying impact in sensitive applications like oncology or forensics.[200]Throughput vs. Cost Trade-offs
DNA sequencing technologies balance throughput—the amount of sequence data produced per unit time or instrument run, often in gigabases (Gb) or terabases (Tb)—against cost, measured per base pair sequenced or per whole human genome equivalent. High-throughput approaches leverage massive parallelism to achieve economies of scale, dramatically reducing marginal costs but frequently compromising on read length, which impacts applications requiring structural variant detection or de novo assembly. Advances in next-generation sequencing (NGS) have decoupled these factors to some extent, with throughput increases outpacing cost reductions via improved chemistry, optics, and flow cell densities, though fundamental engineering limits persist in reagent consumption and error correction.[5] Short-read platforms like Illumina's NovaSeq X series exemplify high-throughput optimization, delivering up to 16 Tb of data per dual flow cell run in approximately 48 hours, enabling over 128 human genomes sequenced per run at costs as low as $200 per 30x coverage genome as of 2024.[201][202] This efficiency stems from sequencing by synthesis with reversible terminators, clustering billions of DNA fragments on a flow cell for simultaneous imaging, yielding per-base costs around $0.01–$0.05. However, read lengths limited to 150–300 base pairs necessitate hybrid mapping strategies and incur higher computational overhead for repetitive genomic regions, where short reads amplify assembly ambiguities.[203] In contrast, long-read technologies trade throughput for extended read lengths to resolve complex structures. Pacific Biosciences' Revio system generates 100–150 Gb of highly accurate HiFi reads (≥Q30 accuracy, 15–20 kb length) per SMRT cell in 12–30 hours, scaling to multiple cells for annual outputs exceeding 100 Tb, but at reagent costs of approximately $11 per Gb, translating to ~$1,000 per human genome.[204][205] This higher per-base expense arises from single-molecule real-time sequencing requiring circular consensus for error correction, limiting parallelism compared to short-read arrays; instrument acquisition costs ~$779,000 further elevate barriers for low-volume users.[206] Oxford Nanopore Technologies' PromethION offers real-time nanopore sequencing with up to 290 Gb per flow cell (R10.4.1 chemistry), supporting ultra-long reads exceeding 10 kb and portability, but initial error rates (5–10%) demand 20–30x coverage for comparable accuracy, pushing costs to ~$1–$1.50 per Gb.[207][208] Flow cell prices range $900–$2,700, with system costs up to $675,000 for high-capacity models, making it suitable for targeted or field applications where immediacy outweighs bulk efficiency.[209]| Platform | Typical Throughput per Run | Approx. Cost per Gb | Avg. Read Length | Key Trade-off |
|---|---|---|---|---|
| Illumina NovaSeq X | 8–16 Tb | $0.01–$0.05 | 150–300 bp | High volume, short reads limit resolution |
| PacBio Revio | 100–150 Gb (per cell) | ~$11 | 15–20 kb (HiFi) | Accurate longs, lower parallelism |
| ONT PromethION | Up to 290 Gb | ~$1–$1.50 | >10 kb | Real-time, higher errors/coverage needs |
Ethical, Legal, and Societal Dimensions
Genetic Privacy and Data Ownership
Genetic data generated through DNA sequencing raises significant privacy concerns due to its uniquely identifiable and immutable nature, distinguishing it from other personal information. Unlike passwords or financial records, genomic sequences can reveal sensitive traits such as disease predispositions, ancestry, and familial relationships, often without explicit consent for secondary uses. In consumer direct-to-consumer (DTC) testing, companies like 23andMe and Ancestry collect saliva samples containing DNA, which users submit voluntarily, but the resulting datasets are stored indefinitely by the firms, creating asymmetries in control. Courts have ruled that once biological material leaves the body, individuals relinquish property rights over it, allowing companies to retain broad commercialization rights over derived data.[211] [212] Ownership disputes center on whether individuals retain sovereignty over their genomic information post-sequencing or if providers assert perpetual claims. DTC firms typically grant users limited access to reports while reserving rights to aggregate, anonymize, and license de-identified data to third parties, including pharmaceutical developers for drug discovery. For instance, 23andMe has partnered with entities like GlaxoSmithKline to share user data for research, justified under terms of service that users often accept without fully grasping implications. Critics argue this commodifies personal biology without equitable benefit-sharing, as companies profit from datasets built on user contributions, yet individuals cannot revoke access or demand deletion of raw sequences once processed. Empirical evidence from privacy audits shows that "anonymized" genetic data remains reidentifiable through cross-referencing with public records or other databases, undermining assurances of detachment from the source individual.[212] [213][214] Data breaches exemplify acute vulnerabilities, as seen in the October 2023 incident at 23andMe, where credential-stuffing attacks—exploiting reused passwords from prior leaks—compromised 6.9 million users' accounts, exposing ancestry reports, genetic relative matches, and self-reported traits but not raw DNA files. The breach stemmed from inadequate multi-factor authentication enforcement, leading to a £2.31 million fine by the UK Information Commissioner's Office in June 2025 for failing to safeguard special category data. In research and clinical sequencing contexts, similar risks persist; for example, hackers could access hospital genomic databases, potentially revealing patient identities via variant patterns unique to 0.1% of the population. These events highlight causal chains where lax security practices amplify harms, including identity theft tailored to genetic profiles or blackmail via inferred health risks.[215][216] [212] Law enforcement access further complicates ownership, as DTC databases enable forensic genealogy to solve cold cases by matching crime scene DNA to relatives' profiles without direct suspect consent. Platforms like GEDmatch allow opt-in uploads, but default privacy settings have led to familial implicature, where innocent relatives' data indirectly aids investigations—over 100 U.S. cases solved by 2019, including the Golden State Killer. Proponents cite public safety benefits, yet detractors note disproportionate impacts on minority groups due to uneven database representation and potential for mission creep into non-criminal surveillance. Companies face subpoenas or voluntary disclosures, with policies varying; 23andMe resists routine sharing but complies under legal compulsion, raising questions of data as a public versus private good.[217] [213] Regulatory frameworks lag behind technological scale, with the U.S. relying on the 2008 Genetic Information Nondiscrimination Act (GINA), which prohibits health insurer or employer misuse but excludes life insurance, long-term care, and data security mandates. No comprehensive federal genetic privacy law exists, leaving governance to state patchwork and company policies; proposed bills like the 2025 Genomic Data Protection Act seek to restrict sales without consent and enhance breach notifications. In contrast, the EU's General Data Protection Regulation (GDPR) classifies genetic data as a "special category" requiring explicit consent, data minimization, and right to erasure, with fines up to 4% of global revenue for violations—evident in enforcement against non-compliant firms. These disparities reflect differing priorities: U.S. emphasis on innovation incentives versus EU focus on individual rights, though both struggle with enforcing ownership in bankruptcy scenarios, as in 23andMe's 2025 filing, where genetic assets risk transfer without user veto.[218] [219] [218]Incidental Findings, Consent, and Return of Results
Incidental findings, also termed secondary findings, refer to the detection of genetic variants during DNA sequencing that are unrelated to the primary clinical or research indication but may have significant health implications, such as pathogenic mutations in genes associated with hereditary cancer syndromes or cardiovascular disorders.[220] These arise particularly in broad-scope analyses like whole-exome or whole-genome sequencing, where up to 1-2% of cases may yield actionable incidental variants depending on the gene panel applied.[221] The American College of Medical Genetics and Genomics (ACMG) maintains a curated list of genes for which laboratories should actively seek and report secondary findings, updated to version 3.2 in 2023, encompassing 59 genes linked to conditions with established interventions like aneurysm repair or lipid-lowering therapies to mitigate risks.[222] Informed consent processes for DNA sequencing must address the potential for incidental findings to ensure participant autonomy, typically involving pre-test counseling that outlines the scope of analysis, risks of discovering variants of uncertain significance (VUS), and options for receiving or declining such results.[223] Guidelines recommend tiered consent models, allowing individuals to select preferences for categories like ACMG-recommended actionable findings versus broader results, as surveys indicate 60-80% of participants prefer learning about treatable conditions while fewer opt for non-actionable ones.[224] In clinical settings, consent forms emphasize the probabilistic nature of findings—e.g., positive predictive values below 50% for some carrier statuses—and potential family implications, requiring genetic counseling to mitigate misunderstanding.[225] Research protocols often lack uniform requirements for incidental finding disclosure, leading to variability; for instance, a 2022 study found that only 40% of genomic research consents explicitly mentioned return policies, prompting calls for standardized templates.[226] Policies on returning incidental findings balance beneficence against harms, with ACMG advocating active reporting of high-penetrance, medically actionable variants in consenting patients to enable preventive measures, as evidenced by cases where early disclosure averted conditions like sudden cardiac death.[227] Empirical studies report minimal long-term psychological distress from such returns; a multi-site analysis of over 1,000 individuals receiving exome/genome results found no clinically significant increases in anxiety or depression scores at 6-12 months post-disclosure, though transient uncertainty from VUS was noted in 10-15% of cases.[228] Critics argue that mandatory broad reporting risks over-medicalization and resource strain, given that only 0.3-0.5% of sequenced genomes yield ACMG-tier findings, and downstream validation costs can exceed $1,000 per case without guaranteed clinical utility.[229] In research contexts, return is generally voluntary and clinician-mediated to confirm pathogenicity via orthogonal methods like Sanger sequencing, with 2023 guidelines emphasizing participant preferences over investigator discretion.[230]Discrimination Risks and Regulatory Overreach Critiques
Genetic discrimination risks arise from the potential misuse of DNA sequencing data, where individuals or groups face differential treatment by insurers, employers, or others due to identified genetic variants predisposing them to diseases. For example, carriers of mutations like BRCA1/2, detectable via sequencing, have historically feared denial of coverage or job opportunities, though empirical cases remain rare post-legislation.[231] [232] The U.S. Genetic Information Nondiscrimination Act (GINA), signed into law on May 21, 2008, with employment provisions effective November 21, 2009, bars health insurers and most employers (those with 15 or more employees) from using genetic information for underwriting or hiring decisions.[233] [234] Despite these safeguards, GINA excludes life, disability, and long-term care insurance; military personnel; and small businesses, creating gaps that could expose sequenced individuals to adverse actions in those domains.[235] Surveys reveal persistent public fears of discrimination, with many unaware of GINA's scope, potentially reducing uptake of sequencing for preventive or research purposes.[236] In population genetics applications of sequencing, risks extend to group-level stigmatization, where variants linked to traits or diseases in specific ancestries could fuel societal biases or policy discriminations, as seen in concerns over identifiable cohorts in biobanks.[237] [238] Proponents of expanded protections argue these fears, even if overstated relative to actual incidents, justify broader nondiscrimination laws, while skeptics note that market incentives and competition among insurers mitigate systemic abuse without further mandates.[239] Critiques of regulatory overreach in DNA sequencing emphasize how agencies like the FDA impose barriers that exceed necessary risk mitigation, stifling innovation and consumer access. The FDA's November 22, 2013, warning letter to 23andMe suspended direct-to-consumer (DTC) health reports for lack of premarket approval, halting services for over two years until phased clearances began in 2015, despite no documented widespread harm from the tests.[240] [241] Critics, including industry analysts, contend this exemplifies precautionary overregulation, prioritizing unproven risks like misinterpreted results over benefits such as early health insights, with false-positive rates in raw DTC data reaching 40% in clinically relevant genes but addressable via improved validation rather than bans.[242] [243] The FDA's May 2024 final rule classifying many laboratory-developed tests (LDTs)—integral to custom sequencing assays—as high-risk medical devices drew rebukes for layering costly compliance (e.g., clinical trials, facility inspections) on labs already under Clinical Laboratory Improvement Amendments (CLIA) oversight, potentially curtailing niche genomic innovations without enhancing accuracy proportionally.[244] [245] A federal district court vacated the rule on March 31, 2025, citing overstepped authority and disruption to the testing ecosystem.[246] Additional measures, such as 2025 restrictions on overseas genetic sample processing, have been lambasted for invoking national security pretexts that inflate costs and delay results, favoring protectionism over evidence-based risks in a globalized field.[247] Such interventions, detractors argue, reflect institutional caution biasing against rapid technological deployment, contrasting with historical under-regulation that enabled sequencing cost drops from $100 million per genome in 2001 to under $1,000 by 2015.[248]Equity, Access, and Innovation Incentives
Advancements in DNA sequencing technologies have significantly reduced costs, enhancing access for broader populations. By 2024, whole genome sequencing costs had fallen to approximately $500 per genome, driven by economies of scale and competitive innovations in next-generation platforms.[202][249] This decline, from millions in the early 2000s to under $1,000 today, has democratized sequencing in high-income settings, enabling routine clinical applications like newborn screening and cancer diagnostics. However, equitable distribution remains uneven, with persistent barriers in low- and middle-income countries where infrastructure, trained personnel, and regulatory frameworks lag.[5][250] Equity concerns arise from underrepresentation of non-European ancestries in genomic databases, comprising over 90% European-origin data in many repositories as of 2022, which skews variant interpretation and polygenic risk scores toward majority populations.[251] This bias perpetuates health disparities, as clinical tools perform poorly for underrepresented groups, such as African or Indigenous ancestries, limiting diagnostic accuracy and personalized medicine benefits. Efforts to address this include targeted recruitment in initiatives like the NIH's All of Us program, aiming for 60% minority participation, yet systemic issues like mistrust from historical abuses and socioeconomic barriers hinder progress.[252][253] Global access disparities are exacerbated by economic and logistical factors, including high out-of-pocket costs, insurance gaps, and rural isolation, disproportionately affecting minorities and underserved communities even in developed nations.[254] In low-resource settings, sequencing uptake is minimal, with fewer than 1% of cases sequenced in many regions during events like the COVID-19 pandemic, underscoring infrastructure deficits.[255] International strategies, such as WHO's genomic surveillance goals for all member states by 2032, seek to mitigate this through capacity-building, but implementation varies due to funding dependencies and geopolitical priorities.[256] Innovation in DNA sequencing is propelled by intellectual property frameworks and market competition, which incentivize R&D investments in high-risk biotechnology. Patents on synthetic methods and sequencing instruments, upheld post the 2013 Supreme Court ruling against natural DNA patentability, protect novel technologies like CRISPR integration and error-correction algorithms, fostering follow-on developments.[257] Competition among platforms, evidenced by market growth from $12.79 billion in 2024 to projected $51.31 billion by 2034, accelerates throughput improvements and cost efficiencies via iterative advancements from firms like Illumina and emerging challengers.[258] Critics argue broad patents can impede downstream research, as studies on gene patents show mixed effects on innovation, with some evidence of reduced follow-on citations in patented genomic regions.[259] Nonetheless, empirical trends indicate that competitive dynamics, rather than monopolistic IP, have been causal in cost trajectories, aligning incentives with broader accessibility gains.[260]Commercial and Economic Aspects
Market Leaders and Technology Platforms
Illumina, Inc. maintains dominance in the next-generation sequencing (NGS) market, commanding approximately 80% share as of 2025 through its sequencing by synthesis (SBS) technology, which relies on reversible dye-terminator nucleotides to generate billions of short reads (typically 100-300 base pairs) per run with high accuracy (Q30+ error rates below 0.1%).[261][262] Key instruments include the NovaSeq 6000 series, capable of outputting up to 20 terabases per flow cell for large-scale genomics projects, and the MiSeq for targeted lower-throughput applications. This platform's entrenched ecosystem, including integrated library preparation and bioinformatics tools, has sustained Illumina's lead despite antitrust scrutiny over acquisitions like Grail.[263] Pacific Biosciences (PacBio) specializes in long-read sequencing via single-molecule real-time (SMRT) technology, using zero-mode waveguides to observe phospholinked fluorescently labeled nucleotides in real time, yielding high-fidelity (HiFi) reads averaging 15-20 kilobases with >99.9% accuracy after circular consensus sequencing. The Revio system, launched in 2023 and scaling to production in 2025, supports up to 1,300 human genomes per year at reduced costs, targeting structural variant detection where short-read methods falter.[264] Oxford Nanopore Technologies (ONT) employs protein nanopores embedded in membranes to measure ionic current disruptions from translocating DNA/RNA strands, enabling real-time, ultra-long reads exceeding 2 megabases and direct epigenetic detection without amplification biases.[80] Devices like the PromethION deliver petabase-scale output, with portability via MinION for field applications, though basecalling error rates (5-10% raw) require computational polishing.[265] MGI Tech, a subsidiary of BGI Genomics, competes with SBS platforms akin to Illumina's but optimized for cost efficiency, particularly in Asia, where it holds significant share through instruments like the DNBSEQ-T7 outputting 12 terabases per run using DNA nanoball technology for higher density. As of 2025, MGI's global expansion challenges Illumina's pricing, with systems priced 30-50% lower, though reagent compatibility and service networks lag in Western markets.[266] Emerging entrants like Ultima Genomics introduce alternative high-throughput approaches, such as multi-cycle SBS on patterned arrays, aiming for sub-$100 genome costs via massive parallelism, but remain niche with limited 2025 adoption.[267]| Platform | Key Technology | Read Length | Strengths | Market Focus |
|---|---|---|---|---|
| Illumina SBS | Reversible terminators | Short (100-300 bp) | High throughput, accuracy | Population genomics, clinical diagnostics[268] |
| PacBio SMRT | Real-time fluorescence | Long (10-20 kb HiFi) | Structural variants, phasing | De novo assembly, rare disease |
| ONT Nanopore | Ionic current sensing | Ultra-long (>100 kb) | Real-time, epigenetics, portability | Infectious disease, metagenomics[80] |
| MGI DNBSEQ | DNA nanoballs + SBS | Short (150-300 bp) | Cost-effective scale | Large cohorts, emerging markets |

