Recent from talks
Nothing was collected or created yet.
Dominance (genetics)
View on WikipediaThis article needs additional citations for verification. (February 2018) |
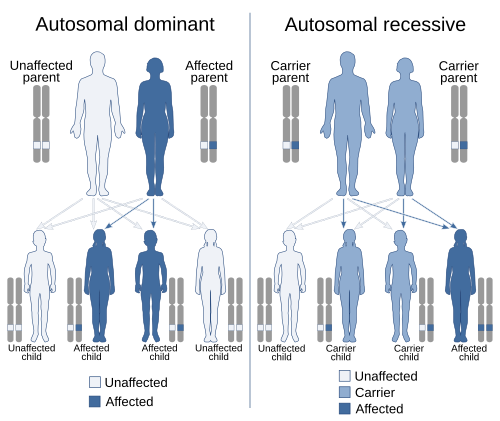
In genetics, dominance is the phenomenon of one variant (allele) of a gene on a chromosome masking or overriding the effect of a different variant of the same gene on the other copy of the chromosome.[1][2] The first variant is termed dominant and the second is called recessive. This state of having two different variants of the same gene on each chromosome is originally caused by a mutation in one of the genes, either new (de novo) or inherited. The terms autosomal dominant or autosomal recessive are used to describe gene variants on non-sex chromosomes (autosomes) and their associated traits, while those on sex chromosomes (allosomes) are termed X-linked dominant, X-linked recessive or Y-linked; these have an inheritance and presentation pattern that depends on the sex of both the parent and the child (see Sex linkage). Since there is only one Y chromosome, Y-linked traits cannot be dominant or recessive. Additionally, there are other forms of dominance, such as incomplete dominance, in which a gene variant has a partial effect compared to when it is present on both chromosomes, and co-dominance, in which different variants on each chromosome both show their associated traits.
Dominance is a key concept in Mendelian inheritance and classical genetics.[3] Letters and Punnett squares are used to demonstrate the principles of dominance in teaching, and the upper-case letters are used to denote dominant alleles and lower-case letters are used for recessive alleles. An often quoted example of dominance is the inheritance of seed shape in peas. Peas may be round, associated with allele R, or wrinkled, associated with allele r. In this case, three combinations of alleles (genotypes) are possible: RR, Rr, and rr. The RR (homozygous) individuals have round peas, and the rr (homozygous) individuals have wrinkled peas. In Rr (heterozygous) individuals, the R allele masks the presence of the r allele, so these individuals also have round peas. Thus, allele R is dominant over allele r, and allele r is recessive to allele R.
Dominance is not inherent to an allele or its traits (phenotype). It is a strictly relative effect between two alleles of a given gene of any function; one allele can be dominant over a second allele of the same gene, recessive to a third, and co-dominant with a fourth. Additionally, one allele may be dominant for one trait but not others. Dominance differs from epistasis, the phenomenon of an allele of one gene masking the effect of alleles of a different gene.
Background
[edit]
Gregor Johann Mendel, "The Father of Genetics", promulgated the idea of dominance in the 1860s. However, it was not widely known until the early twentieth century. Mendel observed that, for a variety of traits of garden peas having to do with the appearance of seeds, seed pods, and plants, there were two discrete phenotypes, such as round versus wrinkled seeds, yellow versus green seeds, red versus white flowers or tall versus short plants.[3] When bred separately, the plants always produced the same phenotypes, generation after generation. However, when lines with different phenotypes were crossed (interbred), one and only one of the parental phenotypes showed up in the offspring (green, round, red, or tall).[3] However, when these hybrid plants were crossed, the offspring plants showed the two original phenotypes, in a characteristic 3:1 ratio, the more common phenotype being that of the parental hybrid plants. Mendel reasoned that each parent in the first cross was a homozygote for different alleles (one parent AA and the other parent aa), that each contributed one allele to the offspring, with the result that all of these hybrids were heterozygotes (Aa), and that one of the two alleles in the hybrid cross dominated expression of the other: A masked a. The final cross between two heterozygotes (Aa X Aa) would produce AA, Aa, and aa offspring in a 1:2:1 genotype ratio with the first two classes showing the (A) phenotype, and the last showing the (a) phenotype, thereby producing the 3:1 phenotype ratio.[3]
Mendel did not use the terms gene, allele, phenotype, genotype, homozygote, and heterozygote, all of which were introduced later.[4][5] He did introduce the notation of capital and lowercase letters for dominant and recessive alleles, respectively, still in use today.
In 1928, British population geneticist Ronald Fisher proposed that dominance acted based on natural selection through the contribution of modifier genes. In 1929, American geneticist Sewall Wright responded by stating that dominance is simply a physiological consequence of metabolic pathways and the relative necessity of the gene involved.[4][6][5]
Types of dominance
[edit]Complete dominance (Mendelian)
[edit]In complete dominance, the effect of one allele in a heterozygous genotype completely masks the effect of the other. The allele that masks are considered dominant to the other allele, and the masked allele is considered recessive.[7]
When we only look at one trait determined by one pair of genes, we call it monohybrid inheritance.[8] If the crossing is done between parents (P-generation, F0-generation) who are homozygote dominant and homozygote recessive, the offspring (F1-generation) will always have the heterozygote genotype and always present the phenotype associated with the dominant gene.[9]

However, if the F1-generation is further crossed with the F1-generation (heterozygote crossed with heterozygote) the offspring (F2-generation) will present the phenotype associated with the dominant gene ¾ times.[8] Although heterozygote monohybrid crossing can result in two phenotype variants, it can result in three genotype variants - homozygote dominant, heterozygote and homozygote recessive, respectively.[10][9]

In dihybrid inheritance we look at the inheritance of two pairs of genes simultaneous. Assuming here that the two pairs of genes are located at non-homologous chromosomes, such that they are not coupled genes (see genetic linkage) but instead inherited independently.[11] Consider now the cross between parents (P-generation) of genotypes homozygote dominant and recessive, respectively. The offspring (F1-generation) will always heterozygous and present the phenotype associated with the dominant allele variant.[11] However, when crossing the F1-generation there are four possible phenotypic possibilities and the phenotypical ratio for the F2-generation will always be 9:3:3:1.[12][11]
Incomplete dominance (non-Mendelian)
[edit]
Incomplete dominance (also called partial dominance, semi-dominance, intermediate inheritance, or occasionally incorrectly co-dominance in reptile genetics[13]) occurs when the phenotype of the heterozygous genotype is distinct from and often intermediate to the phenotypes of the homozygous genotypes. The phenotypic result often appears as a blended form of characteristics in the heterozygous state. For example, the snapdragon flower color is homozygous for either red or white. When the red homozygous flower is paired with the white homozygous flower, the result yields a pink snapdragon flower. The pink snapdragon is the result of incomplete dominance. A similar type of incomplete dominance is found in the four o'clock plant wherein pink color is produced when true-bred parents of white and red flowers are crossed. In quantitative genetics, where phenotypes are measured and treated numerically, if a heterozygote's phenotype is exactly between (numerically) that of the two homozygotes, the phenotype is said to exhibit no dominance at all, i.e. dominance exists only when the heterozygote's phenotype measure lies closer to one homozygote than the other.
When plants of the F1 generation are self-pollinated, the phenotypic and genotypic ratio of the F2 generation will be 1:2:1 (Red:Pink:White).[14]
Co-dominance (non-Mendelian)
[edit]
Co-dominance occurs when the contributions of both alleles are visible in the phenotype and neither allele masks another.
For example, in the ABO blood group system, chemical modifications to a glycoprotein (the H antigen) on the surfaces of blood cells are controlled by three alleles, two of which are co-dominant to each other (IA, IB) and dominant over the recessive i at the ABO locus. The IA and IB alleles produce different modifications. The enzyme coded for by IA adds an N-acetylgalactosamine to a membrane-bound H antigen. The IB enzyme adds a galactose. The i allele produces no modification. Thus the IA and IB alleles are each dominant to i (IAIA and IAi individuals both have type A blood, and IBIB and IBi individuals both have type B blood), but IAIB individuals have both modifications on their blood cells and thus have type AB blood, so the IA and IB alleles are said to be co-dominant.[14]
Another example occurs at the locus for the beta-globin component of hemoglobin, where the three molecular phenotypes of HbA/HbA, HbA/HbS, and HbS/HbS are all distinguishable by protein electrophoresis. (The medical condition produced by the heterozygous genotype is called sickle-cell trait and is a milder condition distinguishable from sickle-cell anemia, thus the alleles show incomplete dominance concerning anemia, see above). For most gene loci at the molecular level, both alleles are expressed co-dominantly, because both are transcribed into RNA.[14]

Co-dominance, where allelic products co-exist in the phenotype, is different from incomplete dominance, where the quantitative interaction of allele products produces an intermediate phenotype. For example, in co-dominance, a red homozygous flower and a white homozygous flower will produce offspring that have red and white spots. When plants of the F1 generation are self-pollinated, the phenotypic and genotypic ratio of the F2 generation will be 1:2:1 (Red:Spotted:White). These ratios are the same as those for incomplete dominance. Again, this classical terminology is inappropriate – in reality, such cases should not be said to exhibit dominance at all.[14]
Relationship to other genetic concepts
[edit]Dominance can be influenced by various genetic interactions and it is essential to evaluate them when determining phenotypic outcomes. Multiple alleles, epistasis, pleiotropic genes, and polygenic characteristics are some factors that might influence the phenotypic outcome.[15]
Multiple alleles
[edit]Although any individual of a diploid organism has at most two different alleles at a given locus, most genes exist in a large number of allelic versions in the population as a whole. This is called polymorphism, and is caused by mutations. Polymorphism can have an effect on the dominance relationship and phenotype, which is observed in the ABO blood group system. The gene responsible for human blood type have three alleles; A, B, and O, and their interactions result in different blood types based on the level of dominance the alleles expresses towards each other.[15][16]
Epistasis
[edit]Epistasis is interactions between multiple alleles at different loci. More specifically, epistasis is when one gene can mask the phenotype of a gene at a completely different locus.[17] Therefore, several genes can influence the phenotype expressed. Epistasis is slightly different from dominance in the fact that dominance is an allele-to-allele interaction at one locus while epistasis is a gene-to-gene interaction at different loci.[18] The dominance relationship between alleles involved in epistatic interactions can influence the observed phenotypic ratios in offspring.[19]
An example of epistasis can be seen in Labrador retriever coat colors. One gene at one locus codes for the color of hair but another gene at a different locus determines if the color is even deposited in the hair.[18][17] Recessive epistasis is seen in this example due to recessive alleles for color desposition masking both the dominant black (B) allele and recessive brown (b) allele at the first locus to express a yellow coat in the Labrador retriever.[18][17] The yellow color comes from no pigment being deposited in the hair shaft.[18]
Other examples of epistasis interactions are dominant epistasis and duplicate recessive epistasis.[18] Each type of epistasis is a modification of the dihyrbid ratio of 9:3:3:1.[17]
Pleiotropic genes
[edit]Pleiotropic genes are genes where one single gene affects two or more characteristics. An example of this concept is Marfan Syndrome which is a mutation of the FBN1 gene. The effects this causes are a person's appearance being tall and long limbed. They can also have Scoliosis, Ectopia Lentis, and larger than normal aortas.[20] Pleiotropy shares a relationship with Epistasis. While pleiotropy represents one single gene, epistasis is multiple genes interacting with one another to cause different traits to arise. it is helpful to recognize how Epistasis could affect viewing pleiotropic genes if different traits arise or mask themselves to varying degrees.[21]
Polygenic characteristics
[edit]Polygenic characteristics are those affected by multiple genes at different loci.[22] These different genes interact in a way to produce a quantitative characteristic, which is a characteristic that presents a wide variety phenotypes, such as height in humans.[22] The greater the number of genes that interact to influence this characteristic, the greater the number of different phenotypes possible due to more possible genotypes.[22] Many more characteristics also appear to be affected by more than one gene located on different loci, including diabetes and some autoimmune diseases.[23]
See also
[edit]References
[edit]- ^ "dominance". Oxford Dictionaries Online. Oxford University Press. Archived from the original on July 18, 2012. Retrieved 14 May 2014.
- ^ "express". Oxford Dictionaries Online. Oxford University Press. Archived from the original on July 18, 2012. Retrieved 14 May 2014.
- ^ a b c d ""Experiments in Plant Hybridization" (1866), by Johann Gregor Mendel | Embryo Project Encyclopedia". embryo.asu.edu. Retrieved 2025-04-27.
- ^ a b Mayo, O. and Bürger, R. 1997. The evolution of dominance: A theory whose time has passed? Archived 2016-03-04 at the Wayback Machine "Biological Reviews", Volume 72, Issue 1, pp. 97–110
- ^ a b "1909: The Word Gene Coined". www.genome.gov. Retrieved 2025-04-27.
- ^ Bourguet, D. 1999. The evolution of dominance Archived 2016-08-29 at the Wayback Machine Heredity, Volume 83, Number 1, pp. 1–4
- ^ Rodríguez-Beltrán, Jerónimo; Sørum, Vidar; Toll-Riera, Macarena; de la Vega, Carmen; Peña-Miller, Rafael; San Millán, Álvaro (2020). "Genetic dominance governs the evolution and spread of mobile genetic elements in bacteria". Proc Natl Acad Sci U S A. 117 (27). United States: United States: National Academy of Sciences: 15755–15762. Bibcode:2020PNAS..11715755R. doi:10.1073/pnas.2001240117. ISSN 0027-8424. PMC 7355013. PMID 32571917.
- ^ a b "18.4: Monohybrid Cross and the Punnett Square". Biology LibreTexts. 2021-10-11. Retrieved 2025-04-27.
- ^ a b "4.2.1: Monohybrid Crosses and Segregation". Biology LibreTexts. 2019-10-01. Retrieved 2025-04-27.
- ^ Trudy, F. C. Mackay; Robert, R. H. Anholt (2022). "Gregor Mendel's legacy in quantitative genetics". PLOS Biology. 20 (7) e3001692. Public Library of Science (PLoS). doi:10.1371/journal.pbio.3001692. ISSN 1544-9173. PMC 9295954. PMID 35852997.
- ^ a b c "6.1: Dihybrid Crosses". Biology LibreTexts. 2016-06-02. Retrieved 2025-04-27.
- ^ Alberts, Bruce; Heald, Rebecca; Hopkin, Karen; Johnson, Alexander; Morgan, David; Roberts, Keith; Walter, Peter (2023). Essential cell biology (Sixth edition.; International student ed.). W.W. Norton & Company. ISBN 978-1-324-03339-4.
- ^ Bulinski, Steven (2016-01-05). "A Crash Course in Reptile Genetics". Reptiles. Living World Media. Archived from the original on 2020-02-04. Retrieved 2023-02-03.
The term co-dominant is often used interchangeably with incomplete dominant, but the two terms have different meanings.
- ^ a b c d Brown, T. A. (2018). Genomes 4 (4th ed.). Milton: Milton: Garland Science. doi:10.1201/9781315226828. ISBN 978-0-8153-4508-4. S2CID 239528980.
{{cite book}}: CS1 maint: publisher location (link) - ^ a b Ingelman-Sundberg, M. (2005). "Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversity". Pharmacogenomics J. 5 (1). United States: United States: Nature Publishing Group: 6–13. doi:10.1038/sj.tpj.6500285. ISSN 1470-269X. PMID 15492763. S2CID 10695794.
- ^ Yamamoto, F; Clausen, H; White, T; Marken, J; Hakomori, S (1990). "Molecular genetic basis of the histo-blood group ABO system". Nature. 345 (6272): 229–233. Bibcode:1990Natur.345..229Y. doi:10.1038/345229a0. PMID 2333095. S2CID 4237562.
- ^ a b c d Urry, Lisa A.; Cain, Michael L.; Wasserman, Steven A.; Minorsky, Peter V.; Orr, Rebecca B. (2021). Campbell Biology (12th ed.). Pearson Education, Inc. pp. 281–282. ISBN 978-0-13-518874-3.
- ^ a b c d e Pierce, Benjamin A. (2024). Genetics: a conceptual approach (Seventh edition digital update ed.). Austin: Macmillan Learning. ISBN 978-1-319-33778-0.
- ^ Phillips, Patrick C (2008). "Epistasis - the essential role of gene interactions in the structure and evolution of genetic systems". Nat Rev Genet. 9 (11). London: London: Nature Publishing Group: 855–867. doi:10.1038/nrg2452. ISSN 1471-0056. PMC 2689140. PMID 18852697.
- ^ Dietz, Harry (1993), Adam, Margaret P.; Feldman, Jerry; Mirzaa, Ghayda M.; Pagon, Roberta A. (eds.), "FBN1-Related Marfan Syndrome", GeneReviews®, Seattle (WA): University of Washington, Seattle, PMID 20301510, retrieved 2025-04-27
- ^ Dwivedi, Sangam L.; Heslop-Harrison, Pat; Amas, Junrey; Ortiz, Rodomiro; Edwards, David (2024). "Epistasis and pleiotropy-induced variation for plant breeding". Plant Biotechnology Journal. 22 (10): 2788–2807. Bibcode:2024PBioJ..22.2788D. doi:10.1111/pbi.14405. ISSN 1467-7652. PMC 11536456. PMID 38875130.
- ^ a b c Pierce, Benjamin A. (2024). Genetics: A conceptual approach (7th ed.). Macmillan Learning.
- ^ Boyle, Evan (2017). "An Expanded View of Complex Traits: From Polygenic to Omnigenic". Cell. 169 (7): 1177–1186. doi:10.1016/j.cell.2017.05.038. PMC 5536862. PMID 28622505.
- "On-line notes for Biology 2250 – Principles of Genetics". Memorial University of Newfoundland.
- Online Mendelian Inheritance in Man (OMIM): Hemoglobin—Beta Locus; HBB - 141900 — Sickle-Cell Anemia
- Online Mendelian Inheritance in Man (OMIM): ABO Glycosyltransferase - 110300 — ABO blood groups
External links
[edit]Dominance (genetics)
View on GrokipediaIntroduction and History
Definition and Basic Principles
In genetics, dominance describes the relationship between alleles—alternative forms of the same gene located at a specific position, or locus, on a chromosome—such that one allele, termed dominant, masks or overrides the expression of another allele, termed recessive, in individuals carrying both (heterozygotes), resulting in a phenotype that matches the homozygous dominant genotype.[8] The genotype refers to an organism's complete set of genes or genetic makeup, including the specific alleles present at each locus, while the phenotype encompasses the observable traits or characteristics arising from the interaction of genotype and environment.[12] A homozygote possesses two identical alleles at a given locus (e.g., AA or aa), whereas a heterozygote has two different alleles (e.g., Aa).[13] This concept is illustrated through a simple monohybrid cross, such as one involving plant height in pea plants, where the allele for tall height (T) is dominant to the allele for short height (t).[14] Consider a cross between two heterozygous individuals (Tt × Tt); the possible offspring genotypes can be predicted using a Punnett square, a diagrammatic tool for determining genotypic outcomes based on parental alleles:| T | t | |
|---|---|---|
| T | TT | Tt |
| t | Tt | tt |

