Recent from talks
Nothing was collected or created yet.
Spectral clustering
View on Wikipedia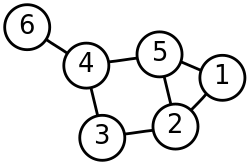

In multivariate statistics, spectral clustering techniques make use of the spectrum (eigenvalues) of the similarity matrix of the data to perform dimensionality reduction before clustering in fewer dimensions. The similarity matrix is provided as an input and consists of a quantitative assessment of the relative similarity of each pair of points in the dataset.
In application to image segmentation, spectral clustering is known as segmentation-based object categorization.
Definitions
[edit]Given an enumerated set of data points, the similarity matrix may be defined as a symmetric matrix , where represents a measure of the similarity between data points with indices and . The general approach to spectral clustering is to use a standard clustering method (there are many such methods, k-means is discussed below) on relevant eigenvectors of a Laplacian matrix of . There are many different ways to define a Laplacian which have different mathematical interpretations, and so the clustering will also have different interpretations. The eigenvectors that are relevant are the ones that correspond to several smallest eigenvalues of the Laplacian except for the smallest eigenvalue which will have a value of 0. For computational efficiency, these eigenvectors are often computed as the eigenvectors corresponding to the largest several eigenvalues of a function of the Laplacian.

Spectral clustering is well known to relate to partitioning of a mass-spring system, where each mass is associated with a data point and each spring stiffness corresponds to a weight of an edge describing a similarity of the two related data points, as in the spring system. Specifically, the classical reference [1] explains that the eigenvalue problem describing transversal vibration modes of a mass-spring system is exactly the same as the eigenvalue problem for the graph Laplacian matrix defined as
- ,
where is the diagonal matrix
and A is the adjacency matrix.
The masses that are tightly connected by the springs in the mass-spring system evidently move together from the equilibrium position in low-frequency vibration modes, so that the components of the eigenvectors corresponding to the smallest eigenvalues of the graph Laplacian can be used for meaningful clustering of the masses. For example, assuming that all the springs and the masses are identical in the 2-dimensional spring system pictured, one would intuitively expect that the loosest connected masses on the right-hand side of the system would move with the largest amplitude and in the opposite direction to the rest of the masses when the system is shaken — and this expectation will be confirmed by analyzing components of the eigenvectors of the graph Laplacian corresponding to the smallest eigenvalues, i.e., the smallest vibration frequencies.
The goal of normalization is making the diagonal entries of the Laplacian matrix to be all unit, also scaling off-diagonal entries correspondingly. In a weighted graph, a vertex may have a large degree because of a small number of connected edges but with large weights just as well as due to a large number of connected edges with unit weights.
A popular normalized spectral clustering technique is the normalized cuts algorithm or Shi–Malik algorithm introduced by Jianbo Shi and Jitendra Malik,[2] commonly used for image segmentation. It partitions points into two sets based on the eigenvector corresponding to the second-smallest eigenvalue of the symmetric normalized Laplacian defined as
The vector is also the eigenvector corresponding to the second-largest eigenvalue of the symmetrically normalized adjacency matrix
The random walk (or left) normalized Laplacian is defined as
and can also be used for spectral clustering. A mathematically equivalent algorithm [3] takes the eigenvector corresponding to the largest eigenvalue of the random walk normalized adjacency matrix .
The eigenvector of the symmetrically normalized Laplacian and the eigenvector of the left normalized Laplacian are related by the identity
Cluster analysis via Spectral Embedding
[edit]Knowing the -by- matrix of selected eigenvectors, mapping — called spectral embedding — of the original data points is performed to a -dimensional vector space using the rows of . Now the analysis is reduced to clustering vectors with components, which may be done in various ways.
In the simplest case , the selected single eigenvector , called the Fiedler vector, corresponds to the second smallest eigenvalue. Using the components of one can place all points whose component in is positive in the set and the rest in , thus bi-partitioning the graph and labeling the data points with two labels. This sign-based approach follows the intuitive explanation of spectral clustering via the mass-spring model — in the low frequency vibration mode that the Fiedler vector represents, one cluster data points identified with mutually strongly connected masses would move together in one direction, while in the complement cluster data points identified with remaining masses would move together in the opposite direction. The algorithm can be used for hierarchical clustering by repeatedly partitioning the subsets in the same fashion.
In the general case , any vector clustering technique can be used, e.g., DBSCAN.
Algorithms
[edit]- Basic Algorithm
- Calculate the Laplacian (or the normalized Laplacian)
- Calculate the first eigenvectors (the eigenvectors corresponding to the smallest eigenvalues of )
- Consider the matrix formed by the first eigenvectors; the -th row defines the features of graph node
- Cluster the graph nodes based on these features (e.g., using k-means clustering)
If the similarity matrix has not already been explicitly constructed, the efficiency of spectral clustering may be improved if the solution to the corresponding eigenvalue problem is performed in a matrix-free fashion (without explicitly manipulating or even computing the similarity matrix), as in the Lanczos algorithm.
For large-sized graphs, the second eigenvalue of the (normalized) graph Laplacian matrix is often ill-conditioned, leading to slow convergence of iterative eigenvalue solvers. Preconditioning is a key technology accelerating the convergence, e.g., in the matrix-free LOBPCG method. Spectral clustering has been successfully applied on large graphs by first identifying their community structure, and then clustering communities.[4]
Spectral clustering is closely related to nonlinear dimensionality reduction, and dimension reduction techniques such as locally-linear embedding can be used to reduce errors from noise or outliers.[5]
Costs
[edit]Denoting the number of the data points by , it is important to estimate the memory footprint and compute time, or number of arithmetic operations (AO) performed, as a function of . No matter the algorithm of the spectral clustering, the two main costly items are the construction of the graph Laplacian and determining its eigenvectors for the spectral embedding. The last step — determining the labels from the -by- matrix of eigenvectors — is typically the least expensive requiring only AO and creating just a -by- vector of the labels in memory.
The need to construct the graph Laplacian is common for all distance- or correlation-based clustering methods. Computing the eigenvectors is specific to spectral clustering only.
Constructing graph Laplacian
[edit]The graph Laplacian can be and commonly is constructed from the adjacency matrix. The construction can be performed matrix-free, i.e., without explicitly forming the matrix of the graph Laplacian and no AO. It can also be performed in-place of the adjacency matrix without increasing the memory footprint. Either way, the costs of constructing the graph Laplacian is essentially determined by the costs of constructing the -by- graph adjacency matrix.
Moreover, a normalized Laplacian has exactly the same eigenvectors as the normalized adjacency matrix, but with the order of the eigenvalues reversed. Thus, instead of computing the eigenvectors corresponding to the smallest eigenvalues of the normalized Laplacian, one can equivalently compute the eigenvectors corresponding to the largest eigenvalues of the normalized adjacency matrix, without even talking about the Laplacian matrix.
Naive constructions of the graph adjacency matrix, e.g., using the RBF kernel, make it dense, thus requiring memory and AO to determine each of the entries of the matrix. Nystrom method[6] can be used to approximate the similarity matrix, but the approximate matrix is not elementwise positive,[7] i.e. cannot be interpreted as a distance-based similarity.
Algorithms to construct the graph adjacency matrix as a sparse matrix are typically based on a nearest neighbor search, which estimate or sample a neighborhood of a given data point for nearest neighbors, and compute non-zero entries of the adjacency matrix by comparing only pairs of the neighbors. The number of the selected nearest neighbors thus determines the number of non-zero entries, and is often fixed so that the memory footprint of the -by- graph adjacency matrix is only , only sequential arithmetic operations are needed to compute the non-zero entries, and the calculations can be trivially run in parallel.
Computing eigenvectors
[edit]The cost of computing the -by- (with ) matrix of selected eigenvectors of the graph Laplacian is normally proportional to the cost of multiplication of the -by- graph Laplacian matrix by a vector, which varies greatly whether the graph Laplacian matrix is dense or sparse. For the dense case the cost thus is . The very commonly cited in the literature cost comes from choosing and is clearly misleading, since, e.g., in a hierarchical spectral clustering as determined by the Fiedler vector.
In the sparse case of the -by- graph Laplacian matrix with non-zero entries, the cost of the matrix-vector product and thus of computing the -by- with matrix of selected eigenvectors is , with the memory footprint also only — both are the optimal low bounds of complexity of clustering data points. Moreover, matrix-free eigenvalue solvers such as LOBPCG can efficiently run in parallel, e.g., on multiple GPUs with distributed memory, resulting not only in high quality clusters, which spectral clustering is famous for, but also top performance.[8]
Software
[edit]Free software implementing spectral clustering is available in large open source projects like scikit-learn[9] using LOBPCG[10] with multigrid preconditioning[11][12] or ARPACK, MLlib for pseudo-eigenvector clustering using the power iteration method,[13] and R.[14]
Relationship with other clustering methods
[edit]The ideas behind spectral clustering may not be immediately obvious. It may be useful to highlight relationships with other methods. In particular, it can be described in the context of kernel clustering methods, which reveals several similarities with other approaches.[15]
Relationship with k-means
[edit]Spectral clustering is closely related to the k-means algorithm, especially in how cluster assignments are ultimately made. Although the two methods differ fundamentally in their initial formulations—spectral clustering being graph-based and k-means being centroid-based—the connection becomes clear when spectral clustering is viewed through the lens of kernel methods.
In particular, weighted kernel k-means provides a key theoretical bridge between the two. Kernel k-means is a generalization of the standard k-means algorithm, where data is implicitly mapped into a high-dimensional feature space through a kernel function, and clustering is performed in that space. Spectral clustering, especially the normalized versions, performs a similar operation by mapping the input data (or graph nodes) to a lower-dimensional space defined by the eigenvectors of the graph Laplacian. These eigenvectors correspond to the solution of a relaxation of the normalized cut or other graph partitioning objectives.
Mathematically, the objective function minimized by spectral clustering can be shown to be equivalent to the objective function of weighted kernel k-means in this transformed space. This was formally established in works such as [16] where they demonstrated that normalized cuts are equivalent to a weighted version of kernel k-means applied to the rows of the normalized Laplacian’s eigenvector matrix.
Because of this equivalence, spectral clustering can be viewed as performing kernel k-means in the eigenspace defined by the graph Laplacian. This theoretical insight has practical implications: the final clustering step in spectral clustering typically involves running the standard k-means algorithm on the rows of the matrix formed by the first k eigenvectors of the Laplacian. These rows can be thought of as embedding each data point or node in a low-dimensional space where the clusters are more well-separated and hence, easier for k-means to detect.
Additionally, multi-level methods have been developed to directly optimize this shared objective function. These methods work by iteratively coarsening the graph to reduce problem size, solving the problem on a coarse graph, and then refining the solution on successively finer graphs. This leads to more efficient optimization for large-scale problems, while still capturing the global structure preserved by the spectral embedding.[17]
Relationship to DBSCAN
[edit]Spectral clustering is also conceptually related to DBSCAN (Density-Based Spatial Clustering of Applications with Noise), particularly in the special case where the spectral method is used to identify connected graph components of a graph. In this trivial case—where the goal is to identify subsets of nodes with no interconnecting edges between them—the spectral method effectively reduces to a connectivity-based clustering approach, much like DBSCAN.[18]
DBSCAN operates by identifying density-connected regions in the input space: points that are reachable from one another via a sequence of neighboring points within a specified radius (ε), and containing a minimum number of points (minPts). The algorithm excels at discovering clusters of arbitrary shape and separating out noise without needing to specify the number of clusters in advance.
In spectral clustering, when the similarity graph is constructed using a hard connectivity criterion (i.e., binary adjacency based on whether two nodes are within a threshold distance), and no normalization is applied to the Laplacian, the resulting eigenstructure of the graph Laplacian directly reveals disconnected components of the graph. This mirrors DBSCAN's ability to isolate density-connected components. The zeroth eigenvectors of the unnormalized Laplacian correspond to these components, with one eigenvector per connected region.
This connection is most apparent when spectral clustering is used not to optimize a soft partition (like minimizing the normalized cut), but to identify exact connected components—which corresponds to the most extreme form of “density-based” clustering, where only directly or transitively connected nodes are grouped together. Therefore, spectral clustering in this regime behaves like a spectral version of DBSCAN, especially in sparse graphs or when constructing ε-neighborhood graphs.
While DBSCAN operates directly in the data space using density estimates, spectral clustering transforms the data into an eigenspace where global structure and connectivity are emphasized. Both methods are non-parametric in spirit, and neither assumes convex cluster shapes, which further supports their conceptual alignment.
Measures to compare clusterings
[edit]Ravi Kannan, Santosh Vempala and Adrian Vetta[19] proposed a bicriteria measure to define the quality of a given clustering. They said that a clustering was an (α, ε)-clustering if the conductance of each cluster (in the clustering) was at least α and the weight of the inter-cluster edges was at most ε fraction of the total weight of all the edges in the graph. They also look at two approximation algorithms in the same paper.
History and related literatures
[edit]Spectral clustering has a long history.[20][21][22][23][24][2][25] Spectral clustering as a machine learning method was popularized by Shi & Malik[2] and Ng, Jordan, & Weiss.[25]
Ideas and network measures related to spectral clustering also play an important role in a number of applications apparently different from clustering problems. For instance, networks with stronger spectral partitions take longer to converge in opinion-updating models used in sociology and economics.[26][27]
See also
[edit]References
[edit]- ^ Demmel, J. "CS267: Notes for Lecture 23, April 9, 1999, Graph Partitioning, Part 2".
- ^ a b c Jianbo Shi and Jitendra Malik, "Normalized Cuts and Image Segmentation", IEEE Transactions on PAMI, Vol. 22, No. 8, Aug 2000.
- ^ Marina Meilă & Jianbo Shi, "Learning Segmentation by Random Walks", Neural Information Processing Systems 13 (NIPS 2000), 2001, pp. 873–879.
- ^ Zare, Habil; Shooshtari, P.; Gupta, A.; Brinkman, R. (2010). "Data reduction for spectral clustering to analyze high throughput flow cytometry data". BMC Bioinformatics. 11 403. doi:10.1186/1471-2105-11-403. PMC 2923634. PMID 20667133.
- ^ Arias-Castro, E.; Chen, G.; Lerman, G. (2011), "Spectral clustering based on local linear approximations.", Electronic Journal of Statistics, 5: 1537–87, arXiv:1001.1323, doi:10.1214/11-ejs651, S2CID 88518155
- ^ Fowlkes, C (2004). "Spectral grouping using the Nystrom method". IEEE Transactions on Pattern Analysis and Machine Intelligence. 26 (2): 214–25. Bibcode:2004ITPAM..26..214F. doi:10.1109/TPAMI.2004.1262185. PMID 15376896. S2CID 2384316.
- ^ Wang, S.; Gittens, A.; Mahoney, M.W. (2019). "Scalable Kernel K-Means Clustering with Nystrom Approximation: Relative-Error Bounds". Journal of Machine Learning Research. 20: 1–49. arXiv:1706.02803.
- ^ Acer, Seher; Boman, Erik G.; Glusa, Christian A.; Rajamanickam, Sivasankaran (2021). "Sphynx: A parallel multi-GPU graph partitioner for distributed-memory systems". Parallel Computing. 106 102769. arXiv:2105.00578. doi:10.1016/j.parco.2021.102769. S2CID 233481603.
- ^ "2.3. Clustering".
- ^ Knyazev, Andrew V. (2003). Boley; Dhillon; Ghosh; Kogan (eds.). Modern preconditioned eigensolvers for spectral image segmentation and graph bisection. Clustering Large Data Sets; Third IEEE International Conference on Data Mining (ICDM 2003) Melbourne, Florida: IEEE Computer Society. pp. 59–62.
- ^ Knyazev, Andrew V. (2006). Multiscale Spectral Image Segmentation Multiscale preconditioning for computing eigenvalues of graph Laplacians in image segmentation. Fast Manifold Learning Workshop, WM Williamburg, VA. doi:10.13140/RG.2.2.35280.02565.
- ^ Knyazev, Andrew V. (2006). Multiscale Spectral Graph Partitioning and Image Segmentation. Workshop on Algorithms for Modern Massive Datasets Stanford University and Yahoo! Research.
- ^ "Clustering - RDD-based API - Spark 3.2.0 Documentation".
- ^ "Kernlab: Kernel-Based Machine Learning Lab". 12 November 2019.
- ^ Filippone, M.; Camastra, F.; Masulli, F.; Rovetta, S. (January 2008). "A survey of kernel and spectral methods for clustering" (PDF). Pattern Recognition. 41 (1): 176–190. Bibcode:2008PatRe..41..176F. doi:10.1016/j.patcog.2007.05.018.
- ^ Dhillon, I.S.; Guan, Y.; Kulis, B. (2004). "Kernel k-means: spectral clustering and normalized cuts" (PDF). Proceedings of the tenth ACM SIGKDD international conference on Knowledge discovery and data mining. pp. 551–6.
- ^ Dhillon, Inderjit; Guan, Yuqiang; Kulis, Brian (November 2007). "Weighted Graph Cuts without Eigenvectors: A Multilevel Approach". IEEE Transactions on Pattern Analysis and Machine Intelligence. 29 (11): 1944–1957. Bibcode:2007ITPAM..29.1944D. CiteSeerX 10.1.1.131.2635. doi:10.1109/tpami.2007.1115. PMID 17848776. S2CID 9402790.
- ^ Schubert, Erich; Hess, Sibylle; Morik, Katharina (2018). The Relationship of DBSCAN to Matrix Factorization and Spectral Clustering (PDF). LWDA. pp. 330–334.
- ^ Kannan, Ravi; Vempala, Santosh; Vetta, Adrian (2004). "On Clusterings : Good, Bad and Spectral". Journal of the ACM. 51 (3): 497–515. doi:10.1145/990308.990313. S2CID 207558562.
- ^ Cheeger, Jeff (1969). "A lower bound for the smallest eigenvalue of the Laplacian". Proceedings of the Princeton Conference in Honor of Professor S. Bochner.
- ^ Donath, William; Hoffman, Alan (1972). "Algorithms for partitioning of graphs and computer logic based on eigenvectors of connections matrices". IBM Technical Disclosure Bulletin.
- ^ Fiedler, Miroslav (1973). "Algebraic connectivity of graphs". Czechoslovak Mathematical Journal. 23 (2): 298–305. Bibcode:1973CzMJ...23..298F. doi:10.21136/CMJ.1973.101168.
- ^ Guattery, Stephen; Miller, Gary L. (1995). "On the performance of spectral graph partitioning methods". Annual ACM-SIAM Symposium on Discrete Algorithms.
- ^ Daniel A. Spielman and Shang-Hua Teng (1996). "Spectral Partitioning Works: Planar graphs and finite element meshes". Annual IEEE Symposium on Foundations of Computer Science.
- ^ a b Ng, Andrew Y.; Jordan, Michael I.; Weiss, Yair (2002). "On spectral clustering: analysis and an algorithm" (PDF). Advances in Neural Information Processing Systems.
- ^ DeMarzo, P. M.; Vayanos, D.; Zwiebel, J. (2003-08-01). "Persuasion Bias, Social Influence, and Unidimensional Opinions". The Quarterly Journal of Economics. 118 (3). Oxford University Press: 909–968. doi:10.1162/00335530360698469. ISSN 0033-5533.
- ^ Golub, Benjamin; Jackson, Matthew O. (2012-07-26). "How Homophily Affects the Speed of Learning and Best-Response Dynamics". The Quarterly Journal of Economics. 127 (3). Oxford University Press (OUP): 1287–1338. doi:10.1093/qje/qjs021. ISSN 0033-5533.
Spectral clustering
View on GrokipediaFundamentals
Overview
Spectral clustering is a nonlinear graph-based unsupervised learning technique that uses the Laplacian matrix and its eigenvectors to perform a kernel-like embedding into a lower-dimensional space, leveraging the eigenvalues and eigenvectors of a similarity matrix to enable the subsequent application of traditional clustering methods such as k-means on the reduced representations.[3][4] This approach treats data points as vertices in a graph, with edges weighted by pairwise similarities, transforming the clustering problem into one of graph partitioning.[2] Intuitively, spectral clustering identifies clusters by relaxing the combinatorial graph cut problem, which seeks to minimize the connections between groups while maintaining balanced partitions, into a continuous optimization solvable via spectral decomposition.[2] This relaxation allows the method to uncover natural groupings based on the connectivity structure of the similarity graph, effectively capturing the underlying manifold or topology of the data without assuming spherical cluster shapes.[4] The basic workflow of spectral clustering involves constructing a similarity graph from the input data, computing the graph Laplacian matrix as a key operator that encodes the graph's spectral properties, extracting the eigenvectors corresponding to the smallest eigenvalues to embed the data into a reduced space, and finally applying k-means clustering to these embeddings to obtain the partitions.[4] Compared to traditional methods like k-means, spectral clustering excels at handling non-convex and irregularly shaped clusters, incorporates rich pairwise similarity information, and is often more robust to noise and initialization sensitivity, leading to superior performance on complex datasets.[4]Graph Construction
In spectral clustering, the data points are represented as vertices in an undirected graph, with edges weighted by measures of similarity between the points to capture their relational structure. The core of this construction is the adjacency matrix , also called the affinity or similarity matrix, where each entry quantifies the similarity between data points and . This matrix forms the basis for subsequent spectral analysis, transforming the clustering problem into one of graph partitioning.[4] A widely used similarity function is the radial basis function (RBF), or Gaussian kernel, given by where denotes the Euclidean distance and is a bandwidth parameter that controls the scale at which similarities are computed—smaller values emphasize local neighborhoods, while larger ones capture broader affinities. This kernel is favored for its smoothness and ability to model non-linear relationships effectively. Other functions, such as polynomial kernels of the form where and , can also be employed to emphasize higher-order interactions, though the Gaussian remains the most common due to its empirical robustness across datasets. The choice of similarity function influences the graph's ability to reveal cluster structure, with parameters like often selected heuristically, such as by setting it to the median pairwise distance in the dataset or via cross-validation to balance under- and over-smoothing.[4][5][2] Several graph types can be constructed from the similarity matrix to balance density, computational efficiency, and representational fidelity. The fully connected graph includes edges between all pairs of vertices with positive similarity weights, resulting in a dense matrix suitable for small datasets but computationally intensive for large ones. To sparsify the graph and reduce complexity, the k-nearest neighbors (k-NN) approach connects each vertex only to its k closest neighbors based on the similarity measure, creating an undirected graph by including mutual connections (i.e., an edge exists if is among 's k-nearest or vice versa); the value of k is typically chosen based on the expected number of points per cluster, often around 5–20. Alternatively, the -neighborhood graph connects vertices if their distance is below a threshold , yielding a sparse structure that thresholds low similarities to zero, with tuned to data density to avoid disconnected components. For domain-specific applications like image segmentation, affinities may incorporate both feature similarities (e.g., color or texture) and spatial proximity, as in for neighboring pixels within radius , where and denote feature and position vectors, respectively.[4][5][2] In high-dimensional data, direct graph construction can suffer from the curse of dimensionality, where distances become less meaningful due to sparse sampling in high-dimensional spaces. To address this, preprocessing steps such as principal component analysis (PCA) are commonly applied to reduce the dimensionality to a manageable level (e.g., 10–50 components retaining 95% variance) before computing similarities, preserving essential structure while mitigating noise and computational costs. This step ensures the graph effectively reflects intrinsic manifolds without excessive sparsity or uniform similarities.[6]Laplacian Matrix
The unnormalized Laplacian matrix of a graph is defined as , where is the affinity (or adjacency) matrix with non-negative entries representing the similarity between nodes and , and is the degree matrix, a diagonal matrix with .[7] This construction positions as the core operator in spectral clustering, transforming the similarity graph into a form amenable to spectral analysis.[7] Key properties of include its symmetry, arising from the symmetric nature of , and positive semi-definiteness, meaning all eigenvalues are non-negative real numbers ordered as .[7] The smallest eigenvalue has multiplicity equal to the number of connected components in the graph, with its corresponding eigenvector being the all-ones vector .[7] These eigenvalues and eigenvectors play a pivotal role in spectral clustering, as the eigenvectors associated with the smallest eigenvalues approximate indicator functions of the graph's connected components, thereby revealing underlying cluster structure.[7] A fundamental characterization of is provided by its quadratic form: for any vector .[7] This expression links the Laplacian directly to graph cuts, quantifying the total squared difference of across weighted edges and thus encoding the smoothness of on the graph; vectors that vary little within clusters but differ between them minimize this form, facilitating effective partitioning.[7]Spectral Embedding
Unnormalized Embedding
In unnormalized spectral embedding, the data points are mapped to a lower-dimensional space using the eigenvectors of the unnormalized Laplacian matrix . Specifically, the eigenvectors corresponding to the smallest non-zero eigenvalues of are computed and stacked as columns to form the embedding matrix , where is the number of data points. The rows of then serve as the coordinates of each data point in , providing a representation that captures the graph's structure for subsequent clustering.[7] The number of clusters is typically selected using the eigengap heuristic, which identifies the value of that maximizes the difference between consecutive eigenvalues. This gap indicates a natural separation in the spectrum, suggesting that the first non-trivial eigenvectors effectively capture the cluster structure while higher ones introduce noise or less relevant variations. The heuristic performs well when clusters are well-separated, as larger eigengaps correspond to more stable partitions under perturbations.[7] Mathematically, this embedding arises from minimizing the Rayleigh quotient associated with , which for a vector is given by where are the edge weights. The eigenvectors corresponding to the smallest eigenvalues minimize this quotient, approximating the solution to the RatioCut objective , where measures the weight of edges crossing between cluster and its complement. By relaxing the discrete cluster indicators to continuous vectors and solving subject to , the columns of (the embedding eigenvectors) provide a relaxed approximation to the optimal partition.[7][7] For the case of two clusters (), the embedding reduces to the second eigenvector of , known as the Fiedler vector, which bipartitions the graph by assigning nodes to clusters based on the sign of their entries (positive to one cluster, negative to the other). This vector, corresponding to the algebraic connectivity , tends to separate the graph into components with minimal edge cuts relative to their sizes, as its nodal values highlight structural bottlenecks.[7]Normalized Embedding
Normalized spectral embedding addresses limitations in unnormalized approaches by incorporating degree normalization, which helps mitigate biases toward clusters of equal size and improves robustness to variations in node degrees. In spectral clustering, two primary normalized Laplacian matrices are used: the symmetric normalized Laplacian and the random walk normalized Laplacian. The symmetric normalized Laplacian is defined as , where is the diagonal degree matrix, is the unnormalized Laplacian, and is the adjacency matrix. This operator is symmetric and positive semi-definite, ensuring its eigenvectors are orthogonal. The random walk normalized Laplacian is given by , which corresponds to the transition matrix of a random walk on the graph and is particularly suited for analyzing connectivity in terms of walk probabilities. For embedding using the symmetric normalized Laplacian , the k smallest eigenvectors are computed to form an matrix , where each row corresponds to a data point's embedding coordinates. To obtain the final normalized embedding, the rows of are then normalized to have unit Euclidean length, yielding coordinates that lie on the unit hypersphere and facilitate subsequent clustering via methods like k-means. This normalization step ensures that the embedding is invariant to scaling differences across nodes, enhancing the separation of clusters with varying densities or sizes. In contrast, embeddings from directly use the eigenvectors without additional row normalization, as the operator already incorporates degree weighting. The benefits of normalized embeddings include greater robustness to uneven degree distributions in the graph, as the normalization accounts for node connectivity strengths, preventing high-degree nodes from dominating the embedding space. The eigenvectors of are orthogonal by construction, providing a stable basis for dimensionality reduction that captures balanced graph cuts. These methods minimize the normalized cut criterion, defined for two partitions and as , where is the sum of edge weights between and , and is the volume of . This objective promotes partitions that balance the cut size relative to cluster volumes, leading to more equitable clusterings compared to unnormalized variants.Algorithms
Shi-Malik Algorithm
The Shi-Malik algorithm, introduced in 2000, represents an early and influential approach to normalized spectral clustering, particularly tailored for image segmentation tasks. It formulates clustering as a graph partitioning problem using the normalized cut (Ncut) criterion, which measures the total dissimilarity between groups relative to the total similarity within groups, thereby avoiding the biases of traditional minimum-cut methods that favor unbalanced partitions. This method leverages a recursive bipartitioning strategy on the graph's normalized Laplacian to achieve multi-way clustering, demonstrating superior performance on segmenting non-convex shapes, such as in hand images, where it successfully identifies coherent regions like the palm and fingers in an 80x100 pixel image with Ncut values below 0.04.[2] The algorithm begins with constructing an affinity graph where nodes represent data points (e.g., pixels), and edge weights encode similarity based on feature differences, such as color and spatial proximity, typically using a Gaussian kernel like for connected pairs within a radius . Next, it computes the random-walk normalized Laplacian , where is the degree matrix and is the affinity matrix, and solves the generalized eigenvalue problem to obtain the eigenvectors corresponding to the smallest eigenvalues. For bipartitioning, the second smallest eigenvector (Fiedler vector) is used to embed the nodes into a 1D space.[2] Clustering proceeds by sorting the nodes according to the Fiedler vector values and selecting the consecutive split point that minimizes the Ncut value, which is approximated as , where is the sum of weights across the partition and measures internal connection strength, to assign nodes to two subsets A and B. The process then recurses on each subgraph, embedding and partitioning subgraphs until a desired number of clusters is reached or stopping criteria are met. These criteria include an Ncut threshold (e.g., 0.04) to ensure meaningful partitions and a stability check, such as verifying that the eigenvector does not exhibit excessive smooth variations via histogram ratios (e.g., below 0.06), preventing over-segmentation.[2] The key innovation lies in this recursive spectral partitioning framework, which combines the global optimization properties of spectral methods with the normalized Laplacian to handle unbalanced cluster sizes and complex topologies, originally motivated by perceptual grouping in vision. In the 2000 publication context, the algorithm was validated on natural images, including a hand segmentation example where it outperformed ratio-cut and min-cut methods by producing balanced, semantically coherent segments on non-convex objects, as visualized in the paper's figures.[2]Pseudocode Outline
Input: Affinity graph G(V, E) with weights W, desired number of clusters k, parameters σ_I, σ_X, r, Ncut_threshold=0.04, stability_threshold=0.06
1. Construct similarity matrix W from features F and positions X using Gaussian affinities within radius r.
2. Compute degree matrix D and normalized Laplacian L_rw = D^{-1}(D - W).
3. While number of current clusters < k:
a. Solve generalized eigenproblem (D - W)y = λ D y for the second smallest eigenvector y_2.
b. Sort the nodes according to the values in y_2.
c. Embed nodes using y_2 (1D).
d. Find the split point among consecutive segments in the sorted order that minimizes Ncut(A, B) to bipartition into A and B.
e. Compute Ncut(A, B).
f. If Ncut(A, B) > Ncut_threshold or stability check fails (e.g., histogram ratio > stability_threshold), stop recursion on this branch.
g. Else, recurse on subgraphs G[A] and G[B].
Output: Cluster assignments for k partitions.
Input: Affinity graph G(V, E) with weights W, desired number of clusters k, parameters σ_I, σ_X, r, Ncut_threshold=0.04, stability_threshold=0.06
1. Construct similarity matrix W from features F and positions X using Gaussian affinities within radius r.
2. Compute degree matrix D and normalized Laplacian L_rw = D^{-1}(D - W).
3. While number of current clusters < k:
a. Solve generalized eigenproblem (D - W)y = λ D y for the second smallest eigenvector y_2.
b. Sort the nodes according to the values in y_2.
c. Embed nodes using y_2 (1D).
d. Find the split point among consecutive segments in the sorted order that minimizes Ncut(A, B) to bipartition into A and B.
e. Compute Ncut(A, B).
f. If Ncut(A, B) > Ncut_threshold or stability check fails (e.g., histogram ratio > stability_threshold), stop recursion on this branch.
g. Else, recurse on subgraphs G[A] and G[B].
Output: Cluster assignments for k partitions.
Ng-Jordan-Weiss Algorithm
The Ng-Jordan-Weiss algorithm is a widely used normalized spectral clustering method that embeds data points into a low-dimensional space via eigenvectors of the graph Laplacian and then applies k-means clustering to obtain the final partition.[3] Introduced in 2001, it emphasizes simplicity and effectiveness for identifying clusters in non-convex data structures.[3] The approach assumes the number of clusters is known and proceeds in five main steps, outlined as follows: construct the similarity graph and affinity matrix ; compute the symmetric normalized Laplacian ; extract the eigenvectors corresponding to the smallest eigenvalues; normalize the rows of the eigenvector matrix; and apply k-means to the normalized rows. First, construct a similarity graph from the data points, where vertices represent the points and edge weights form the affinity matrix based on pairwise similarities, such as a Gaussian kernel for points .[3] Second, compute the symmetric normalized Laplacian matrix , where is the diagonal degree matrix with (as detailed in the Normalized Embedding section).[3] Third, find the eigenvectors corresponding to the smallest eigenvalues of , and form the matrix with these as columns.[3] Fourth, normalize each row of to have unit length: for the -th row , replace it with . This row normalization step ensures that points with high degrees in the graph do not dominate the embedding, promoting balanced contributions from all data points.[3] Fifth, apply the k-means algorithm directly to the rows of the normalized to assign each point to one of clusters.[3] The algorithm naturally handles graphs with disconnected components, as the multiplicity of the zero eigenvalue in equals the number of connected components, allowing the eigenvectors to capture the intrinsic cluster structure without additional preprocessing.[3] Empirically, the method demonstrates strong performance on toy datasets such as the Two Moons, concentric Circles, interleaved circles, and joined rings, as well as real-world examples like author affiliation clustering from NIPS proceedings, often outperforming direct k-means by effectively separating non-linearly separable groups.[3] Its advantages include conceptual simplicity—implementable in a few lines of code—and scalability for datasets with moderate size (up to thousands of points), owing to the use of standard linear algebra routines for eigenvector computation.[3]Computational Complexity
Graph Laplacian Construction
The construction of the graph Laplacian in spectral clustering begins with the adjacency matrix , which encodes pairwise similarities between data points in -dimensional space, typically derived from a similarity kernel applied to Euclidean distances. For a full (dense) graph, computing the pairwise Euclidean distances requires time, as each of the pairs involves a distance calculation across dimensions.[8] The resulting dense adjacency matrix and Laplacian (where is the degree matrix) each occupy space, making storage a significant concern for moderate-sized datasets.[8] To address scalability, sparse representations are often employed, such as -nearest neighbor (k-NN) graphs, where only the most similar neighbors per point are retained. This reduces space complexity to , assuming , while the time for constructing the sparse via exact k-NN remains in the brute-force case but can drop to using approximate nearest neighbor methods like tree-based indexing (e.g., KD-trees) in low dimensions, though high dimensions may require other approximations due to the curse of dimensionality.[9] The Laplacian is then formed similarly, inheriting the sparsity, though degree computations add negligible overhead.[10] The primary bottleneck in Laplacian construction is the pairwise similarity computation, which dominates for high-dimensional or large-scale data due to its quadratic scaling.[11] Mitigation strategies include subsampling the data or using landmark (anchor) points to approximate the full similarity structure, reducing the effective computation to interactions with a smaller set of anchors and achieving linear or subquadratic time in .[11] For datasets with , full dense graphs become computationally infeasible, exceeding typical memory limits and requiring hours or days on standard hardware for distance calculations alone.[12] This threshold underscores the need for sparse or approximate approaches, as the resulting Laplacian's size directly impacts subsequent embedding steps.[10]Eigenvector Computation
The computation of eigenvectors is a central step in spectral clustering, where the goal is to obtain the k smallest eigenvectors of the graph Laplacian matrix L to form the spectral embedding. For dense matrices, exact eigendecomposition via methods such as the QR algorithm incurs a time complexity of O(n^3), where n is the number of data points, as it requires decomposing the full n × n matrix. This approach becomes prohibitive for moderate to large n due to its cubic scaling. In practice, iterative methods like Lanczos or Arnoldi are preferred for extracting only the k smallest eigenvectors, achieving a complexity of O(n^2 k) on dense matrices, since each iteration involves O(n^2) operations for matrix-vector multiplications and roughly O(k) iterations are needed for convergence to the partial spectrum. For sparse Laplacians with O(n k) nonzeros (e.g., from k-NN graphs), iterative methods like Lanczos achieve approximately O(n k^2) time complexity for computing k eigenvectors, enabling scalability to n ≈ 10^5 or larger on standard hardware.[13] Significant challenges arise in eigenvector computation, particularly with the Laplacian's properties. The matrix L can be ill-conditioned in noisy graphs, where perturbations from extraneous edges lead to small eigengaps between the k-th and (k+1)-th eigenvalues, causing slow convergence of iterative solvers like Lanczos. Additionally, the need to compute only the partial spectrum (the k smallest eigenvectors) rather than the full set is essential for efficiency, as full decomposition is unnecessary and computationally wasteful for clustering tasks. These issues are exacerbated in real-world data with noise, where eigenvector estimates may deviate substantially from ideal cluster indicators. In terms of practical feasibility, exact eigenvector computation via dense methods is viable on standard desktop hardware for n up to approximately 10^3, taking seconds to minutes depending on k and implementation. Beyond this scale, approximations such as Nyström methods or randomized techniques are required to maintain tractability without full eigendecomposition. Memory requirements also pose a bottleneck, as storing the dense Laplacian demands O(n^2) space, which for n=10^3 uses about 8 MB for double-precision entries but escalates quadratically to gigabytes for larger n.Implementations
Available Software
Spectral clustering implementations are available in several popular programming languages and frameworks, enabling researchers and practitioners to apply the method to various datasets. In Python, the scikit-learn library provides theSpectralClustering class, which implements the Ng-Jordan-Weiss (NJW) algorithm among other variants, using ARPACK as the default eigensolver for computing the required eigenvectors.[14] Key parameters include affinity='rbf' for radial basis function kernels to construct the similarity matrix and n_neighbors to build k-nearest neighbors graphs when the full affinity matrix is computationally expensive.[14] As of 2025, the spectralcluster Python package provides an additional re-implementation of key algorithms, complementing scikit-learn. Few from-scratch repositories exist, often for educational purposes, and NetworkX provides graph datasets suitable for testing.[15]
In R, the kernlab package offers the specc function for spectral clustering, which embeds data into the eigenspace of an affinity matrix derived from kernel functions.[16] For graph-based applications, the igraph package, extended by the rSpectral add-on, supports spectral clustering on network structures through functions like spectral_igraph_membership, which leverage the leading eigenvectors for partitioning.[17]
MATLAB's Statistics and Machine Learning Toolbox includes the built-in spectralcluster function, which performs spectral clustering on data matrices and relies on the eigs function to solve the generalized eigenvalue problem for the graph Laplacian.[18] This allows custom implementations by combining eigs with k-means on the embedded coordinates, supporting normalized and unnormalized variants.[19]
For large-scale and distributed computing, Apache Spark's MLlib provides Power Iteration Clustering (PIC), a scalable approximation to spectral clustering that approximates top eigenvectors via power iterations, suitable for massive graphs.[20] In recent years, PyTorch Geometric has facilitated GPU-accelerated implementations of spectral clustering through optimized sparse matrix operations and eigensolvers, with enhancements in versions from the early 2020s enabling faster processing on NVIDIA hardware for graph neural network-related tasks.
Optimization Techniques
Spectral clustering implementations often face challenges in scalability and robustness due to the high computational cost of eigendecomposition on large graphs and sensitivity to parameters or data perturbations. Optimization techniques address these by introducing approximations, automated parameter tuning, efficient computation strategies, and noise mitigation methods, enabling application to datasets with millions of points while preserving clustering quality. These approaches build on the core spectral framework by reducing matrix sizes, leveraging randomization, or enhancing graph construction, thereby improving both efficiency and practical utility. Approximation methods such as the Nyström technique provide a low-rank approximation to the affinity matrix, which is particularly useful when the matrix exhibits low intrinsic dimensionality. In the Nyström method, a subset of landmark points is selected to form a smaller submatrix, whose eigendecomposition approximates the full matrix's spectrum, reducing the complexity from O(n^3) to O(m^2 n) where m << n is the number of landmarks. This approach was introduced for spectral clustering by Fowlkes et al., who demonstrated its effectiveness in recovering manifold structures in image segmentation tasks. Landmark-based subsampling further refines this by strategically selecting representative points to construct a reduced graph representation, allowing the embedding to be computed on the landmarks and then propagated to all points via Nyström extension or barycentric coordinates. Chen and Cai's landmark spectral clustering method, for instance, selects landmarks via k-means on a subset and uses a low-rank approximation to achieve linear scalability, showing improved performance on large-scale datasets like handwritten digits. Parameter selection is crucial for spectral clustering's success, as choices like the kernel bandwidth σ in radial basis function (RBF) graphs and the number of clusters k directly influence the graph's connectivity and the eigengap in the Laplacian spectrum. For σ, cross-validation techniques evaluate multiple bandwidth values by assessing clustering stability or downstream task performance, such as silhouette scores on held-out data, to select the value that best captures local manifold structure without over-smoothing. von Luxburg recommends this heuristic for RBF kernels to ensure the graph is connected yet discriminative. For k, the eigengap heuristic identifies the optimal number by selecting the largest difference between consecutive eigenvalues of the Laplacian, as this gap often corresponds to the separation between cluster subspaces. This method, formalized in early spectral clustering analyses, reliably estimates k in settings with well-separated clusters, avoiding exhaustive search. Scalability tricks leverage sparse structures and randomized algorithms to bypass full eigendecomposition. Sparse approximations construct the affinity matrix using only nearest neighbors or low-degree graphs, minimizing storage and computation while approximating the full dense matrix's spectrum through techniques like random binning features that map data to a sparse embedding space. Yan et al.'s fast approximate spectral clustering employs such sparsification via local scaling and subset selection, achieving near-linear time on graphs with thousands of nodes. For eigenvector computation, randomized SVD accelerates the extraction of top-k eigenvectors by projecting the matrix onto a low-dimensional random sketch, followed by a deterministic SVD on the reduced space, with error bounds preserving the spectral properties essential for k-means post-processing. Wu et al. (2018) use random binning features to approximate the affinity matrix in a low-dimensional space, enabling scalable spectral clustering to high-dimensional data with speedups of over 100x on benchmark datasets without significant accuracy loss.[21] Handling noise and outliers in spectral clustering requires robust kernels or preprocessing to prevent perturbations from distorting the graph Laplacian. Robust kernels, such as those incorporating Student's t-distribution instead of Gaussian, downweight outlier influences in affinity computation, maintaining cluster separability under contamination levels up to 20%. Li et al.'s noise-robust spectral clustering uses a warping model to map noisy data to a cleaner space before embedding, improving accuracy on synthetic datasets with Gaussian noise. Preprocessing steps like outlier detection via density-based methods (e.g., identifying points with low degree in the k-nearest neighbor graph) or robust PCA to denoise the feature space further enhance resilience, ensuring the subsequent spectral steps focus on true manifold structure.Theoretical Connections
Relation to Graph Partitioning
Spectral clustering originates from graph partitioning problems, where the goal is to divide a graph's vertices into subsets that minimize the number of edges crossing between subsets, known as cuts. The classic min-cut problem, which seeks to bipartition a graph while minimizing the cut size, is NP-hard, prompting the development of approximation methods. Spectral clustering addresses this by relaxing the discrete optimization into a continuous one through eigenvector minimization of the graph Laplacian, providing a computationally tractable surrogate that often yields high-quality partitions. A key formulation in this context is the normalized cut (NCut), introduced by Shi and Malik, which balances partition quality with size constraints to avoid degenerate solutions like isolating single vertices. The NCut for a bipartition into sets and is defined as: where is the sum of edge weights between and , and is the total connection from subset to the graph . This criterion minimizes the normalized cut value while encouraging balanced partitions, and its relaxation leads to solving for the second smallest eigenvector of the normalized Laplacian, approximating the optimal discrete cut. For multi-way partitioning into subsets , the generalized NCut extends to , relaxed by using the first eigenvectors to embed vertices into a lower-dimensional space for subsequent clustering.[2] The theoretical foundation linking spectral methods to graph partitioning is bolstered by the Cheeger inequality, which connects the second smallest eigenvalue of the normalized Laplacian to the graph's conductance (a measure of expansion). Specifically, the inequality states , where is the Cheeger constant, the minimum conductance over all cuts. This provides a spectral guarantee that small implies the existence of a sparse cut, justifying the use of the Fiedler vector (the eigenvector corresponding to ) for identifying balanced partitions with provable approximation quality relative to the optimal cut. For multi-way extensions, higher-order Cheeger inequalities relate the -th eigenvalue gaps to multi-way conductance, enabling spectral approximations for -way cuts.[22]Approximation Guarantees
Spectral clustering algorithms provide theoretical approximation guarantees under specific conditions, particularly when relating to graph partitioning objectives like the balanced k-cut problem. These guarantees often stem from the relaxation of discrete optimization problems into continuous eigenvector computations, followed by rounding steps such as k-means. For instance, a recursive variant of spectral clustering, closely related to the Ng-Jordan-Weiss (NJW) algorithm, achieves a polynomial-time approximation for the balanced k-cut, delivering a clustering with conductance within an O(log n) factor of the optimal value, where n is the number of nodes. This result holds in the worst case using a bicriteria measure that balances cut quality and balance constraints, ensuring the algorithm finds partitions close to optimal when strong clusterings exist.[23] Perturbation theory plays a crucial role in establishing the stability of spectral clustering under noise in the graph or similarity matrix. The eigenvectors of the graph Laplacian are sensitive to perturbations, but matrix perturbation bounds, such as those from the Davis-Kahan theorem, quantify this stability: for an ε-perturbation in the matrix norm, the angular error between the true and perturbed eigensubspaces is bounded by O(ε / δ), where δ is the eigengap between the k-th and (k+1)-th eigenvalues. In the context of spectral clustering, this implies that if the graph is a noisy version of an ideal cluster graph with a sufficiently large eigengap, the embedded points remain close to the ideal cluster indicators, leading to accurate recovery after rounding. This O(ε) error scaling (assuming constant gap) ensures robustness to small amounts of graph noise, such as edge perturbations in real-world data.[24][5] In stochastic block models (SBMs), which generate graphs with planted communities, spectral clustering exhibits sharp recovery thresholds for community detection. Basic spectral methods, using the leading eigenvectors of the expected adjacency or Laplacian matrix, achieve weak recovery—correlating better than random with the true communities—precisely when the signal-to-noise ratio (SNR) exceeds 1, defined as (a - b)^2 / (k (a + (k-1)b)), where a and b are within- and between-community edge probabilities, and k is the number of communities. This threshold matches the information-theoretic detectability limit in the sparse regime (average degree Θ(log n)), enabling exact recovery with high probability above the Kesten-Stigum bound. These guarantees highlight spectral clustering's efficiency in assortative SBMs, outperforming random guessing below the threshold.[25] Recent theoretical advances have extended these guarantees to dynamic and more general settings. For example, dynamic spectral clustering algorithms provide provable approximation guarantees for evolving graphs, achieving amortized update times of O(1) and sublinear query times under mild cluster structure assumptions. Additionally, tighter analyses have shown that spectral clustering can perform well under weaker conditions, and using fewer than k eigenvectors may yield better empirical results in practice.[26][27] Despite these strengths, spectral clustering lacks strong approximation guarantees for arbitrary data distributions, as its performance relies on assumptions like underlying manifold structures or low-dimensional embeddings that align with the Laplacian's spectrum. Without such structure—e.g., in high-dimensional or non-graph-representable data—the method may fail to approximate optimal clusterings, reverting to heuristic behavior without provable bounds. This limitation underscores the need for domain-specific validations beyond theoretical settings.[24]Comparisons with Other Methods
Similarities to K-Means
Spectral clustering and k-means clustering share a fundamental procedural similarity in their final clustering step, where both algorithms apply k-means to a low-dimensional representation of the data. In spectral clustering, the data points are first embedded into a lower-dimensional space using the eigenvectors of the graph Laplacian, which captures the graph's structure based on pairwise similarities. This embedding serves as the input to k-means, mirroring the direct application of k-means to the original data points in the standard algorithm.[1] A key analogy exists between spectral clustering and kernel k-means, where the spectral embedding can be viewed as an implicit form of kernel principal component analysis (PCA). Specifically, when the similarity graph is constructed using a radial basis function (RBF) kernel, the resulting affinity matrix functions as a kernel matrix, allowing spectral clustering to perform nonlinear transformations akin to kernel methods. This connection highlights how spectral clustering extends k-means by incorporating kernel-induced geometries without explicitly computing them in the feature space.[1][28] Spectral clustering exhibits particularly close behavior to k-means when the data forms well-separated spherical clusters, in which case the spectral approach effectively reduces to standard k-means. Both methods aim to minimize within-cluster variance, with spectral clustering achieving this through the optimization of graph cuts that align with Euclidean distances in such scenarios. In the Ng-Jordan-Weiss algorithm, for instance, the final k-means step on the embedded points reinforces this overlap for compact, isotropic clusters.[1] Insights from the early 2000s further elucidate these similarities, demonstrating equivalence between normalized spectral clustering and kernel k-means under specific normalizations of the graph Laplacian. For example, using the random walk Laplacian ensures that the spectral objective corresponds to a weighted kernel k-means formulation, unifying the two in terms of their relaxation to eigenvector problems. This equivalence holds particularly for positive semi-definite kernels and balanced cluster weights, providing theoretical grounding for their shared efficacy on certain data distributions.[1][28]Differences from DBSCAN
Spectral clustering and DBSCAN represent fundamentally different paradigms in unsupervised clustering: the former relies on constructing a similarity graph to model global data manifold structure and relaxes the graph partitioning problem using eigenvectors of the graph Laplacian, while the latter is a density-based method that discovers clusters by identifying local density peaks and expanding regions around core points without presupposing the number of clusters .[7] This graph-relaxation approach in spectral clustering enables it to capture nonlinear structures effectively, assuming the data lies on a low-dimensional manifold represented by the graph, whereas DBSCAN operates directly in the original feature space to detect arbitrary-shaped clusters as high-density areas separated by low-density noise.[24][29] One key strength of spectral clustering over DBSCAN lies in its suitability for non-Euclidean data manifolds and scenarios requiring roughly equal-sized clusters, as the spectral embedding facilitates partitioning into balanced components via subsequent k-means application on the eigenvectors.[7] In contrast, DBSCAN excels at handling clusters of arbitrary shapes and robustly identifying outliers as noise, making it preferable for datasets with irregular geometries or contaminated points where global graph assumptions may falter. However, spectral clustering's performance is highly sensitive to the choice of the affinity matrix parameter , which scales the Gaussian kernel and can distort the graph if misspecified, and it implicitly assumes balanced cluster sizes, potentially leading to suboptimal partitions in imbalanced data.[7] DBSCAN, while free from specifying , requires careful tuning of the neighborhood radius and minimum points to define density, rendering it vulnerable to failures in datasets with varying local densities.[29] Empirically, spectral clustering often underperforms on datasets with heterogeneous densities, where its global graph structure may overlook local variations, as seen in multi-shape benchmarks where adjusted Rand index (ARI) scores for spectral clustering lag behind DBSCAN's 0.96.[30] Conversely, DBSCAN struggles with elongated or intertwined clusters, such as spirals, achieving lower ARI (0.33 ± 0.17) compared to spectral clustering's 0.78 ± 0.38, due to its reliance on uniform density thresholds that fail to capture extended structures.[30] In synthetic two-circle datasets, DBSCAN tends to merge components or oversegment based on (e.g., 25 vs. 26), while spectral clustering separates them but remains noise-sensitive without density integration.[31] These trade-offs highlight spectral clustering's affinity for manifold-like data versus DBSCAN's prowess in noise-resilient, shape-flexible partitioning.[30]Evaluation Metrics
Internal Validation
Internal validation of spectral clustering assesses the quality of the resulting partitions using only the input data and the clustering output, without relying on external ground truth labels. This approach is particularly useful for unsupervised scenarios where labeled data is unavailable, focusing on intrinsic properties such as cluster cohesion, separation, and compactness derived from the spectral embedding or graph structure. Common metrics include the eigengap, silhouette score applied to embeddings, properties of Laplacian eigenvalues, and modularity for graph-based inputs. The eigengap measures cluster separability by examining the difference between the (k+1)-th and k-th smallest eigenvalues of the Laplacian matrix, denoted as , where k is the number of clusters. A larger eigengap indicates well-separated clusters, as it reflects a spectral gap in the graph's connectivity, making the choice of k more robust. This metric is rooted in the theoretical analysis of spectral graph partitioning, where the eigengap bounds the expansion properties of the graph cuts. The silhouette score, when computed on the low-dimensional embeddings obtained from the eigenvectors corresponding to the smallest k eigenvalues, evaluates the cohesion within clusters and separation between them in the embedded k-dimensional space. For each data point, the score is calculated as , where is the average distance to other points in the same cluster and is the minimum average distance to points in a neighboring cluster; the overall score is the mean across all points, with values closer to 1 indicating stronger structure. This adaptation leverages the geometric interpretation of spectral embeddings, providing a way to quantify how well the nonlinear manifold structure is preserved in the reduced space. The trace of the smallest k Laplacian eigenvalues serves as an indicator of cluster compactness, representing the total quadratic form , where L is the Laplacian matrix. Lower trace values suggest tighter clusters, as they correspond to smoother functions over the graph that align with the partition, minimizing the cut size normalized by cluster volumes. This metric directly ties to the Rayleigh quotient optimization in spectral clustering, offering a global measure of how well the embedding captures intra-cluster similarities. For graph-structured data, modularity adapted to spectral partitions quantifies the strength of division by comparing intra-cluster edge densities to a null model, given by , where A is the adjacency matrix, m is the total number of edges, d_i and d_j are degrees, and is 1 if nodes i and j are in the same cluster. Values of Q near 1 indicate strong community structure, while adaptations for spectral methods involve evaluating this on the k-means assignment in the embedding space to assess how well the spectral approximation recovers modular communities. This metric, originally from network analysis, has been shown to correlate with spectral clustering performance on benchmark graphs like stochastic block models.External Validation
External validation assesses the quality of spectral clustering results by comparing predicted clusters to ground truth labels available in benchmark datasets, providing an objective measure of accuracy through extrinsic metrics that quantify agreement between partitions.[32] These metrics are particularly useful for evaluating spectral clustering's ability to recover underlying data structures in labeled scenarios, such as when testing on standard datasets like Iris or MNIST, where performance is compared against baselines like k-means.[33] The Adjusted Rand Index (ARI) is a widely used metric for external validation, measuring the similarity between two clusterings by counting pairs of samples that are assigned in the same or different clusters in each, adjusted for chance agreements to account for random labelings.[34] ARI values range from -1 (complete disagreement) to 1 (perfect agreement), with 0 indicating clustering no better than random; it is robust to variations in cluster sizes and has been applied to spectral clustering evaluations on datasets like Iris, where it highlights improvements over traditional methods.[35] Normalized Mutual Information (NMI) provides an entropy-based measure of shared information between predicted and true clusters, defined aswhere is the mutual information between clusterings and , and denotes entropy.[36] NMI is normalized to the range [0, 1], with 1 signifying identical clusterings and values near 0 indicating independence; in spectral clustering benchmarks on MNIST, NMI scores demonstrate strong alignment with digit labels.[37] The Fowlkes-Mallows Index (FMI) evaluates clustering similarity via the geometric mean of precision and recall on pairwise agreements between predicted and true labels, emphasizing the proportion of correctly paired samples within and across clusters.[38] Ranging from 0 (no similarity) to 1 (perfect match), FMI is sensitive to cluster overlap and has been used alongside ARI and NMI in spectral clustering assessments on datasets like Iris, where it confirms high recovery rates for non-convex structures.[39] In representative benchmarks on datasets such as Iris and MNIST, spectral clustering often outperforms alternatives, establishing its efficacy for complex data geometries while relying on these metrics for rigorous comparison.


































