Recent from talks
Nothing was collected or created yet.
Translocase
View on WikipediaTranslocase is a general term for a protein that assists in moving another molecule, usually across a cell membrane. These enzymes catalyze the movement of ions or molecules across membranes or their separation within membranes. The reaction is designated as a transfer from “side 1” to “side 2” because the designations “in” and “out”, which had previously been used, can be ambiguous.[1] Translocases are the most common secretion system in Gram positive bacteria.
It is also a historical term for the protein now called elongation factor G, due to its function in moving the transfer RNA (tRNA) and messenger RNA (mRNA) through the ribosome.
History
[edit]The enzyme classification and nomenclature list was first approved by the International Union of Biochemistry in 1961. Six enzyme classes had been recognized based on the type of chemical reaction catalyzed, including oxidoreductases (EC 1), transferases (EC 2), hydrolases (EC 3), lyases (EC 4), isomerases (EC 5) and ligases (EC 6). However, it became apparent that none of these could describe the important group of enzymes that catalyse the movement of ions or molecules across membranes or their separation within membranes. Several of these involve the hydrolysis of ATP and had been previously classified as ATPases (EC 3.6.3.-), although the hydrolytic reaction is not their primary function. In August 2018, the International Union of Biochemistry and Molecular Biology classified these enzymes under a new enzyme class (EC) of translocases (EC 7).[2]
Mechanism of catalysis
[edit]The reaction most translocases catalyse is:
- AX + Bside 1|| = A + X + || Bside 2[3]
A clear example of an enzyme that follows this scheme is H+-transporting two-sector ATPase:
- ATP + H2O + 4 H+side 1 = ADP + phosphate + 4 H+side 2

A) ATP-ADP translocase: protein responsible for the 1: 1 exchange of intramitochondrial ATP for ADP produced in the cytoplasm. B) Phosphate translocase: the transport of H2PO4- together with a proton are produced by symport H2PO4-/H+
This ATPase carries out the dephosphorylation of ATP into ADP while it transports H+ to the other side of the membrane.[4]
However, other enzymes that also fall into this category do not follow the same reaction scheme. This is the case of ascorbate ferrireductase:
- ascorbateside 1 + Fe(III)side 2 = monodehydroascorbateside 1 + Fe(II)side 2
In which the enzyme only transports an electron in the catalysation of an oxidoreductase reaction between a molecule and an inorganic cation located on different sides of the membrane.[5]
Function
[edit]The basic function, as already mentioned (see: Translocase § Definition), is to "catalyse the movement of ions or molecules across membranes or their separation within membranes". This form of membrane transport is classified under active membrane transport, an energy-requiring process of pumping molecules and ions across membranes against a concentration gradient.[6]
Translocases biological importance relies primarily on their critical function, in the way that they provide movement across the cell's membrane in many cellular processes that are substantial, such as:
- Oxidative phosphorylation
- ADP/ATP translocase (ANT) imports adenosine diphosphate ADP from the cytosol and exports ATP from the mitochondrial matrix, which are key transport steps for oxidative phosphorylation in eukaryotic organisms. ADP from the cytosol is transported back into the mitochondrion for ATP synthesis and the synthesised ATP, produced from oxidative phosphorylation, is exported out of the mitochondrion for use in the cytosol, providing the cells with its main energy currency.[7]

- Protein import into mitochondria
- Hundreds of proteins encoded by the nucleus are required for mitochondrial metabolism, growth, division, and partitioning to daughter cells, and all of these proteins must be imported into the organelle.[8] Translocase of the outer membrane (TOM) and translocase of the inner membrane (TIM) mediate the import of proteins into the mitochondrion. The translocase of the outer membrane (TOM) sorts proteins via several mechanisms either directly to the outer membrane, the intermembrane space, or the translocase of the inner membrane (TIM). Then, generally, the TIM23 machinery mediates protein translocation into the matrix and the TIM22 machinery mediates insertion into the inner membrane.[9]
- Fatty acids import into mitochondria (Carnitine Shuttle System)
- Carnitine-acylcarnitine translocase (CACT) catalyzes both unidirectional transport of carnitine and carnitine/acylcarnitine exchange in the inner mitochondrial membrane, allowing the import of long-chain fatty acids into the mitochondria where they are oxidized by the β-oxidation pathway.[10] The mitochondrial membrane is impermeable to long-chain fatty acids, hence the need for this translocation.[11]
Classification
[edit]The enzyme subclasses designate the types of components that are being transferred, and the sub-subclasses indicate the reaction processes that provide the driving force for the translocation.[12]
Source:[13]
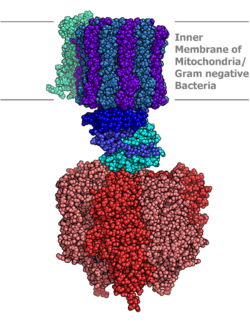
This subclass contains translocases that catalyze the translocation of hydrons.[14] Based on the reaction they are linked to, EC 7.1 can be further classified into:
- EC 7.1.1 Hydron translocation or charge separation linked to oxidoreductase reactions
- EC 7.1.2 Hydron translocation linked to the hydrolysis of a nucleoside triphosphate
- EC 7.1.3 Hydron translocation linked to the hydrolysis of diphosphate
An important translocase contained in this group is ATP synthase, also known as EC 7.1.2.2.

EC 7.2 Catalysing the translocation of inorganic cations and their chelates
[edit]This subclass contains translocases that transfer inorganic cations (metal cations).[15] Based on the reaction they're linked to, EC 7.2 can be further classified into:
- EC 7.2.1 Translocation of inorganic cations linked to oxidoreductase reactions
- EC 7.2.2 Translocation of inorganic cations linked to the hydrolysis of a nucleoside triphosphate
- EC 7.2.4 Translocation of inorganic cations linked to decarboxylation
An important translocase contained in this group is Na+/K+ pump, also known as EC 7.2.2.13.
EC 7.3 Catalysing the translocation of inorganic anions
[edit]This subclass contains translocases that transfer inorganic cations anions. Subclasses are based on the reaction processes that provide the driving force for the translocation. At present only one subclass is represented: EC 7.3.2 Translocation of inorganic anions linked to the hydrolysis of a nucleoside triphosphate.[16]
- 7.3.2.1 ABC-type phosphate transporter
- The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. A bacterial enzyme that interacts with an extracytoplasmic substrate binding protein and mediates the high affinity uptake of phosphate anions. Unlike P-type ATPases, it does not undergo phosphorylation during the transport process.[17]
- ATP + H2O + phosphate [phosphate - binding protein][side 1] = ADP + phosphate + phosphate [side 2] + [phosphate - binding protein][side 1]
- 7.3.2.2 ABC-type phosphonate transporter
- The enzyme, found in bacteria, interacts with an extracytoplasmic substrate binding protein and mediates the import of phosphonate and organophosphate anions.[18]
- ATP + H2O + phosphonate [phosphonate-binding protein][side 1] = ADP + phosphate + phosphonate [side 2] + [phosphonate- binding protein][side 1]
- 7.3.2.3 ABC-type sulfate transporter
- The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. The enzyme from Escherichia coli can interact with either of two periplasmic binding proteins and mediates the high affinity uptake of sulfate and thiosulfate. May also be involved in the uptake of selenite, selenate and possibly molybdate. Does not undergo phosphorylation during the transport.[19]
- ATP + H2O + sulfate [sulfate - binding protein] [side 1] = ADP + phosphate + sulfate [side 2] + [sulfate - binding protein][side 1]
- 7.3.2.4 ABC-type nitrate transporter
- The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. The enzyme, found in bacteria, interacts with an extracytoplasmic substrate binding protein and mediates the import of nitrate, nitrite, and cyanate.[20]
- ATP + H2O + nitrate [nitrate - binding protein][side 1] = ADP + phosphate + nitrate [side 2] + [nitrate - binding protein][side 1]
- 7.3.2.5 ABC-type molybdate transporter
- The expected taxonomic range for this enzyme is: Archaea, Eukaryota, Bacteria. The enzyme, found in bacteria, interacts with an extracytoplasmic substrate binding protein and mediates the high-affinity import of molybdate and tungstate. Does not undergo phosphorylation during the transport process.[21]
- ATP + H2O + molybdate [molybdate - binding protein][side 1] = ADP + phosphate + molybdate [side 2] + [molybdate - binding protein][side 1]
- 7.3.2.6 ABC-type tungstate transporter
- The expected taxonomic range for this enzyme is: Archaea, Bacteria. The enzyme, characterized from the archaeon Pyrococcus furiosus, the Gram-positive bacterium Peptoclostridium acidaminophilum (Eubacterium acidaminophilum) and the Gram-negative bacterium Campylobacter jejuni, interacts with an extracytoplasmic substrate binding protein and mediates the import of tungstate into the cell for incorporation into tungsten-dependent enzymes.[22]
- ATP + H2O + tungstate [tungstate - binding protein][side 1] = ADP + phosphate + tungstate [side 2] + [tungstate - binding protein][side 1]
EC 7.4 Catalysing the translocation of amino acids and peptides
[edit]Subclasses are based on the reaction processes that provide the driving force for the translocation. At present there is only one subclass: EC 7.4.2 Translocation of amino acids and peptides linked to the hydrolysis of a nucleoside triphosphate.[23]
- 7.4.2.1 ABC-type polar-amino-acid transporter
- The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. The enzyme, found in bacteria, interacts with an extracytoplasmic substrate binding protein and mediates the import of polar amino acids. This entry comprises bacterial enzymes that import Histidine, Arginine, Lysine, Glutamine, Glutamate, Aspartate, ornithine, octopine and nopaline.[24]
- ATP + H2O + polar amino acid [polar amino acid-binding protein][side 1] = ADP + phosphate + polar amino acid [side 2] + [polar amino acid-binding protein][side1]
- 7.4.2.2 ABC-type nonpolar-amino-acid transporter
- The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. The enzyme, found in bacteria, interacts with an extracytoplasmic substrate binding protein. This entry comprises enzymes that import Leucine, Isoleucie and Valine.[25]
- ATP + H2O + non polar amino acid [non polar amino acid - binding protein][side 1] = ADP + phosphate + non polar amino acid [side 2] + [non polar amino acid - binding protein][side 1]
- 7.4.2.3 ABC-type mitochondrial protein-transporting ATPase
- The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. A non-phosphorylated, non-ABC (ATP-binding cassette) ATPase involved in the transport of proteins or preproteins into mitochondria using the TIM (Translocase of the Inner Membrane) protein complex. TIM is the protein transport machinery of the mitochondrial inner membrane that contains three essential TIM proteins: Tim17 and Tim23 are thought to build a preprotein translocation channel while Tim44 interacts transiently with the matrix heat-shock protein Hsp70 to form an ATP-driven import motor.[26]
- ATP + H2O + mitochondrial protein [side 1] = ADP + phosphate + mitochondrial protein [side 2]
- 7.4.2.4 ABC-type chloroplast protein-transporting ATPase
- The enzyme appears in viruses and cellular organisms. Involved in the transport of proteins or preproteins into chloroplast stroma (several ATPases may participate in this process).[27]
- ATP + H2O + chloroplast protein [side 1] = ADP + phosphate + chloroplast protein [side 2]
- 7.4.2.5 ABC-type protein transporter
- The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. This entry stands for a family of bacterial enzymes that are dedicated to the secretion of one or several closely related proteins belonging to the toxin, protease and lipase families. Examples from Gram-negative bacteria include α-hemolysin, cyclolysin, colicin V and siderophores, while examples from Gram-positive bacteria include bacteriocin, subtilin, competence factor and pediocin.[28]
- ATP + H2O + protein [side 1] = ADP + phosphate + protein [side 2]
- 7.4.2.6 ABC-type oligopeptide transporter
- A bacterial enzyme that interacts with an extracytoplasmic substrate binding protein and mediates the import of oligopeptides of varying nature. The binding protein determines the specificity of the system. Does not undergo phosphorylation during the transport process.[29]
- ATP + H2O + oligopeptide [oligopeptide - binding protein][side 1] = ADP + phosphate + oligopeptide [side 2] + [oligopeptide - binding protein][side 1]
- 7.4.2.7 ABC-type alpha-factor-pheromone transporter
- The enzyme appears in viruses and cellular organisms characterized by the presence of two similar ATP-binding domains/proteins and two integral membrane domains/proteins. Does not undergo phosphorylation during the transport process. A yeast enzyme that exports the α-factor sex pheromone.[30]
- ATP + H2O + alpha factor [side 1] = ADP + phosphate + alpha factor [side 2]
- 7.4.2.8 ABC-type protein-secreting ATPase
- The expected taxonomic range for this enzyme is: Archaea, Bacteria. A non-phosphorylated, non-ABC (ATP-binding cassette) ATPase that is involved in protein transport.[31]
- ATP + H2O + cellular protein [side 1] = ADP + phosphate + cellular protein [side 2]
- 7.4.2.9 ABC-type dipeptide transporter
- The enzyme appears in viruses and cellular organisms. ATP-binding cassette (ABC) type transporter, characterized by the presence of two similar ATP-binding domains/proteins and two integral membrane domains/proteins. A bacterial enzyme that interacts with an extracytoplasmic substrate binding protein and mediates the uptake of dipeptides and tripeptides.[32]
- ATP + H2O + dipeptide [dipeptide - binding protein][side 1] = ADP + phosphate + [side 2] + [dipeptide - binding protein][side 1]
- 7.4.2.10 ABC-type glutathione transporter
- A prokaryotic ATP-binding cassette (ABC) type transporter, characterized by the presence of two similar ATP-binding domains/proteins and two integral membrane domains/proteins. The enzyme from the bacterium Escherichia coli is a heterotrimeric complex that interacts with an extracytoplasmic substrate binding protein to mediate the uptake of glutathione.[33]
- ATP + H2O glutathione [glutathione - binding protein][side 1] = ADP + phosphate + glutathione [side 2] + [glutathione - binding protein][side 1]
- 7.4.2.11 ABC-type methionine transporter
- A bacterial enzyme that interacts with an extracytoplasmic substrate binding protein and functions to import methionine.[34][35]
- (1) ATP + H2O + L-methionine [methionine - binding protein][side 1] = ADP + phosphate + L-methionine [side 2] + [methionine - binding protein][side 1]
- (2) ATP + H2O + D-methionine [methionine - binding protein][side 1] = ADP + phosphate + D-methionine [side 2] + [methionine - binding protein][side 1]
- 7.4.2.12 ABC-type cystine transporter
- A bacterial enzyme that interacts with an extracytoplasmic substrate binding protein and mediates the high affinity import of trace cystine. The enzyme from Escherichia coli K-12 can import both isomers of cystine and a variety of related molecules including djenkolate, lanthionine, diaminopimelate and homocystine.[36]
- (1) ATP + H2O + L-cystine [cystine - binding protein][side 1] = ADP + phosphate + L-cystine [side 2] + [cystine - binding protein][side 1]
- (2) ATP + H2O + D-cystine [cystine - binding protein][side 1] = ADP + phosphate + D-cystine [side 2] + [cystine - binding protein][side 1]
EC 7.5 Catalysing the translocation of carbohydrates and their derivatives
[edit]- EC 7.5.2 Linked to the hydrolysis of a nucleoside triphosphate
EC 7.6 Catalysing the translocation of other compounds
[edit]- EC 7.6.2 Linked to the hydrolysis of a nucleoside triphosphate
Examples
[edit]- ornithine translocase (SLC25A15), associated with ornithine translocase deficiency.
- carnitine-acylcarnitine translocase (SLC25A20), associated with carnitine-acylcarnitine translocase deficiency.
- Translocase of outer mitochondrial membrane 40 (TOMM40), a protein encoded by the TOMM40 gene, whose alleles differentially impact the risk for Alzheimer's disease
References
[edit]- ^ "EC class 7". ExplorEnz - The Enzyme Database. Retrieved 24 October 2019.
- ^ Tipton K. "Translocases (EC 7): A new EC Class". ExplorEnz - The Enzyme Database. Retrieved 20 October 2019.
- ^ Tipton K, McDonald A (2018). "A Brief Guide to Enzyme Nomenclature and Classification" (PDF).
- ^ "ExplorEnz: EC 7.1.2.2". www.enzyme-database.org. Retrieved 2019-10-24.
- ^ "BRENDA - Information on EC 7.2.1.3 - ascorbate ferrireductase (transmembrane)". www.brenda-enzymes.org. Retrieved 2019-10-24.
- ^ "Active Transport". CK-12 Foundation. Retrieved 2019-10-25.
- ^ Kunji ER, Aleksandrova A, King MS, Majd H, Ashton VL, Cerson E, et al. (October 2016). "The transport mechanism of the mitochondrial ADP/ATP carrier" (PDF). Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. Channels and transporters in cell metabolism. 1863 (10): 2379–93. doi:10.1016/j.bbamcr.2016.03.015. PMID 27001633.
- ^ Ryan KR, Jensen RE (November 1995). "Protein translocation across mitochondrial membranes: what a long, strange trip it is". Cell. 83 (4): 517–9. doi:10.1016/0092-8674(95)90089-6. PMID 7585952.
- ^ Koehler CM (June 2000). "Protein translocation pathways of the mitochondrion". FEBS Letters. Birmingham Issue. 476 (1–2): 27–31. Bibcode:2000FEBSL.476...27K. doi:10.1016/S0014-5793(00)01664-1. PMID 10878244.
- ^ Palmieri F (2008-07-01). "Diseases caused by defects of mitochondrial carriers: a review". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 15th European Bioenergetics Conference 2008. 1777 (7–8): 564–78. doi:10.1016/j.bbabio.2008.03.008. PMID 18406340.
- ^ "Fatty Acids -- Transport and Regeneration". library.med.utah.edu. Retrieved 2019-10-26.
- ^ "EC class 7". ExplorEnz - The Enzyme Database. Retrieved 24 October 2019.
- ^ Hydron is a generic term that includes protons (1H+), deuterons (2H+) and tritons (3H+).
- ^ "EC 7.1 - Catalysing the translocation of hydrons". IntEnz (Integrated relational Enzyme database). Retrieved 24 October 2019.
- ^ "EC 7.2 - Catalysing the translocation of inorganic cations". IntEnz (Integrated relational Enzyme database). Retrieved 24 October 2019.
- ^ "IntEnz - EC 7.3". www.ebi.ac.uk. Retrieved 2019-10-26.
- ^ "BRENDA - Information on EC 7.3.2.1 - ABC-type phosphate transporter". www.brenda-enzymes.org. Retrieved 2019-10-26.
- ^ "BRENDA - Information on EC 7.3.2.2 - ABC-type phosphonate transporter". www.brenda-enzymes.org. Retrieved 2019-10-26.
- ^ "BRENDA - Information on EC 7.3.2.3 - ABC-type sulfate transporter". www.brenda-enzymes.org. Retrieved 2019-10-26.
- ^ "BRENDA - Information on EC 7.3.2.4 - ABC-type nitrate transporter". www.brenda-enzymes.org. Retrieved 2019-10-26.
- ^ "BRENDA - Information on EC 7.3.2.5 - ABC-type molybdate transporter". www.brenda-enzymes.org. Retrieved 2019-10-26.
- ^ "BRENDA - Information on EC 7.3.2.6 - ABC-type tungstate transporter". www.brenda-enzymes.org. Retrieved 2019-10-26.
- ^ "IntEnz - EC 7.4". www.ebi.ac.uk. Retrieved 2019-10-26.
- ^ "BRENDA - Information on EC 7.4.2.1 - ABC-type polar-amino-acid transporter". www.brenda-enzymes.org. Retrieved 2019-10-26.
- ^ "BRENDA - Information on EC 7.4.2.2 - ABC-type nonpolar-amino-acid transporter". www.brenda-enzymes.org. Retrieved 2019-10-26.
- ^ "BRENDA - Information on EC 7.4.2.3 - mitochondrial protein-transporting ATPase". www.brenda-enzymes.org. Retrieved 2019-10-26.
- ^ "BRENDA - Information on EC 7.4.2.4 - chloroplast protein-transporting ATPase". www.brenda-enzymes.org. Retrieved 2019-10-26.
- ^ "BRENDA - Information on EC 7.4.2.5 - ABC-type protein transporter". www.brenda-enzymes.org. Retrieved 2019-10-26.
- ^ "BRENDA - Information on EC 7.4.2.6 - ABC-type oligopeptide transporter". www.brenda-enzymes.org. Retrieved 2019-10-26.
- ^ "BRENDA - Information on EC 7.4.2.7 - ABC-type alpha-factor-pheromone transporter". www.brenda-enzymes.org. Retrieved 2019-10-26.
- ^ "BRENDA - Information on EC 7.4.2.8 - protein-secreting ATPase". www.brenda-enzymes.org. Retrieved 2019-10-26.
- ^ "BRENDA - Information on EC 7.4.2.9 - ABC-type dipeptide transporter". www.brenda-enzymes.org. Retrieved 2019-10-26.
- ^ "Rhea - Annotated reactions database". www.rhea-db.org. Retrieved 2019-10-26.
- ^ "Rhea - Annotated reactions database". www.rhea-db.org. Retrieved 2019-10-26.
- ^ "Rhea - Annotated reactions database". www.rhea-db.org. Retrieved 2019-10-26.
- ^ "IntEnz - EC 7.4.2.12". www.ebi.ac.uk. Retrieved 2019-10-26.
Translocase
View on GrokipediaOverview
Definition
A translocase is a general term for a protein that facilitates the movement of other molecules, typically across biological membranes, without altering their chemical structure. These proteins play a crucial role in cellular transport processes by enabling the directed relocation of ions, metabolites, or macromolecules from one side of a membrane to the other, often harnessing energy sources such as ATP hydrolysis or electrochemical gradients to drive the translocation.[1] In enzyme nomenclature, translocases were formally recognized as the seventh class (EC 7) by the International Union of Biochemistry and Molecular Biology (IUBMB) in August 2018, distinguishing them from the previous six classes established in 1961. This classification addresses enzymes whose primary catalytic function is the translocation of substrates across membranes or their separation within membranes, rather than traditional chemical transformations like oxidation or hydrolysis. Previously, many such enzymes were erroneously grouped under ATPases (EC 3.6.3) where energy coupling was secondary to the translocation event.[1] Unlike broader categories of membrane transporters, which encompass both enzymatic and non-enzymatic mechanisms for solute movement, translocases in the EC 7 class are specifically enzymatic, with translocation defined as the catalyzed reaction. The general reaction schema for EC 7 enzymes is represented as the transfer of a solute from "side 1" of the membrane to "side 2," avoiding directional ambiguities like "in" or "out," and frequently coupled to energy input from nucleoside triphosphate hydrolysis, oxidoreductase activities, or other driving forces.[1]Biological Importance
Translocases are essential for establishing and maintaining electrochemical gradients across biological membranes, which underpin critical cellular processes including signaling and energy production. These enzymes, classified under EC 7, actively translocate ions or metabolites against concentration gradients, generating proton motive forces that power ATP synthesis via oxidative phosphorylation in mitochondria and facilitate secondary transport in plasma membranes. For example, the adenine nucleotide translocase (ANT) exchanges cytosolic ADP for mitochondrial ATP, directly coupling respiratory chain activity to cellular energy demands and ensuring efficient distribution of high-energy phosphates.[6] Similarly, ion-pumping translocases such as the Na⁺/K⁺-ATPase maintain resting membrane potentials vital for neuronal excitability and muscle contraction by counter-transporting sodium and potassium ions.[7] In addition to ion and metabolite handling, translocases are indispensable for protein import into organelles like mitochondria and the endoplasmic reticulum (ER), thereby preventing the cytosolic accumulation of unfolded or mislocalized precursor proteins that could trigger proteotoxic stress. Mitochondrial translocases, including the TOM complex in the outer membrane and TIM complexes in the inner membrane, recognize nuclear-encoded preproteins via targeting signals and thread them across lipid bilayers in an ATP- and membrane potential-dependent manner.[8] This process ensures timely delivery to matrix, inner membrane, or intermembrane space destinations, averting aggregation of immature polypeptides in the cytosol. In the ER, the Sec61 translocon serves as the primary conduit for co- and post-translational protein translocation, inserting nascent polypeptides into the lumen or membrane while coordinating with chaperones to maintain folding competence and avoid cytosolic exposure of hydrophobic domains.[9] Proteomics studies indicate that approximately 99% of human mitochondrial proteins are nuclear-encoded and imported via translocases, despite the organelle encoding only 13 proteins via its own genome.[10] The broad biological importance of translocases is further evidenced by their deep evolutionary conservation across prokaryotes and eukaryotes, reflecting their pivotal role in membrane biogenesis and compartmentalization since early cellular evolution. Prokaryotic translocase systems, such as SecYEG (analogous to eukaryotic Sec61), Tat, and YidC/Oxa1, originated to insert proteins into bacterial plasma membranes and have been repurposed in eukaryotic organelles following endosymbiosis.[11] This conservation extends to mitochondrial biogenesis, where translocases like SAM50 derive from bacterial β-barrel assembly machinery, enabling the integration of outer membrane proteins essential for organelle integrity.[12] Such evolutionary persistence underscores how translocases have been indispensable for adapting membrane dynamics to diverse physiological demands, from prokaryotic energy transduction to eukaryotic multicellularity.Historical Development
Early Discoveries
The discovery of ion translocases began in the mid-20th century with investigations into active transport mechanisms across cell membranes. In 1957, Jens Christian Skou identified Na⁺/K⁺-ATPase in crab nerve membranes, an enzyme that hydrolyzes ATP to drive the exchange of sodium and potassium ions against their concentration gradients, establishing the concept of ATP-dependent ion translocation.[13] This finding, initially classified under EC 3.6.3 for ATPases, highlighted the role of membrane-bound proteins in solute movement and laid foundational insights into energy-coupled transport, though its translocase nature was not fully appreciated until later re-evaluations. Building on these biochemical assays, studies in the 1950s and 1960s focused on mitochondrial energy coupling, culminating in Peter Mitchell's chemiosmotic theory proposed in 1961. Mitchell posited that proton translocation across the inner mitochondrial membrane generates an electrochemical gradient that drives ATP synthesis, integrating observations of anion and cation movements with respiratory chain activity. This theory connected early empirical data on mitochondrial solute exchanges, including the ADP/ATP carrier, to broader principles of membrane potential-driven transport. Subsequent 1960s experiments by Klingenberg and Vignais demonstrated specific ADP/ATP exchange across the inner mitochondrial membrane, revealing an antiporter mechanism that imports ADP for oxidative phosphorylation and exports ATP, further supporting Mitchell's framework through inhibitor studies with atractyloside.[14] In the 1970s, reconstitution experiments using proteoliposomes provided direct evidence of translocase-dependent solute movement. Researchers solubilized and reincorporated mitochondrial membrane proteins into artificial lipid vesicles, demonstrating ATP-driven ADP/ATP exchange that mimicked native transport kinetics and was sensitive to specific inhibitors like carboxyatractyloside. These assays confirmed the carrier's role as a functional translocase, isolating its activity from other membrane components and quantifying exchange rates under controlled gradients. The 1980s brought discoveries in protein translocation, particularly through genetic screens in bacteria. In 1981, Oliver and Beckwith isolated Escherichia coli mutants defective in exporting multiple secreted proteins, identifying the Sec pathway components such as SecA, an ATPase motor that powers translocation across the cytoplasmic membrane.[15] This system, involving the SecYEG translocon, recognized the need for dedicated protein translocases to thread unfolded polypeptides through membranes, expanding the scope of translocase mechanisms beyond ions to macromolecules. In the 1990s, advances in eukaryotic protein import revealed the translocase of the outer mitochondrial membrane (TOM) complex, identified through biochemical fractionation and genetic studies in yeast, serving as the entry gate for mitochondrial precursor proteins. Similarly, the translocase of the inner membrane (TIM) complexes, particularly TIM23, were characterized for driving protein insertion using the proton motive force, building on earlier bacterial models.[3]Establishment of EC 7
In 2018, the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB) established a new enzyme class, EC 7, dedicated to translocases, recognizing their distinct catalytic role in facilitating the movement of ions or molecules across membranes or within membranes.[1] This decision addressed the limitations of the existing six enzyme classes (EC 1–6), which were insufficient to classify these membrane transport proteins accurately, as their primary function—translocation—had been overshadowed by secondary reactions like hydrolysis in prior assignments.[16] Enzymes previously categorized under EC 3 (hydrolases), particularly the ATPases in subclass EC 3.6.3, were transferred to EC 7 to better reflect their translocation activity as the main enzymatic reaction.[1] The rationale for this reclassification stemmed from the need to prioritize the translocation mechanism over ancillary processes, such as ATP hydrolysis, which had led to misplacements in earlier nomenclature systems.[17] Reactions in EC 7 are now denoted using a standardized "side 1" to "side 2" notation, where substrates move from one side of a membrane (or compartment) to another, with subclasses (e.g., EC 7.1–7.6) delineating the translocated species and sub-subclasses specifying the driving forces, such as redox reactions or nucleotide hydrolysis.[1] This shift enhanced the precision of enzyme classification, aligning it more closely with functional biochemistry and supporting advances in membrane protein research.[16] The establishment of EC 7 prompted updates to major enzyme databases, with the initial transfers integrated into ExplorEnz (the IUBMB's primary nomenclature resource) and other resources following the 2018 decision. Ongoing additions continue to expand the class as new translocases are characterized, ensuring the nomenclature remains dynamic.[18] The key publication outlining this change is the IUBMB's "Brief Guide to Enzyme Nomenclature" (2018 edition), which formalized the new class and its structural framework.[16]Classification
EC 7.1: Hydron Translocation
EC 7.1 comprises enzymes that catalyze the translocation of hydrons (protons, H⁺) across biological membranes, typically from one side (side 1) to the other (side 2), generating or utilizing electrochemical gradients essential for cellular energy processes. This subclass is part of the broader translocases (EC 7) introduced by the International Union of Biochemistry and Molecular Biology (IUBMB) in 2018 to classify membrane-bound enzymes previously scattered across other EC classes. The translocation is frequently coupled to exergonic reactions, such as the hydrolysis of nucleoside triphosphates like ATP or redox reactions in electron transport systems, enabling vectorial proton movement that drives ATP synthesis or maintains membrane potential.[1][19] The enzymes in EC 7.1 are subdivided based on the coupled reaction: EC 7.1.1 for those linked to oxidoreductase activities, EC 7.1.2 for nucleoside triphosphate hydrolysis, and EC 7.1.3 for diphosphate hydrolysis. As of May 2023, this subclass includes 18 distinct entries, though the exact count evolves with ongoing classifications of respiratory and photosynthetic complexes.[18] Prominent examples include EC 7.1.1.2 (NADH:ubiquinone reductase, H⁺-translocating), known as the proton-translocating NADH dehydrogenase or Complex I of the mitochondrial electron transport chain, which transfers electrons from NADH to ubiquinone while pumping four protons per NADH oxidized. Another key enzyme is EC 7.1.2.2 (H⁺-transporting two-sector ATPase), the F₀F₁-ATP synthase found in mitochondria, bacteria, and chloroplasts, which reversibly couples proton influx to ATP synthesis.[20][18] These enzymes are vital for pH homeostasis, as they regulate intracellular and compartmental proton concentrations to prevent acidification or alkalization that could disrupt enzymatic activities. In energy metabolism, they contribute to ATP production by establishing proton motive force; for instance, in oxidative phosphorylation, proton translocation by EC 7.1.1 enzymes builds the gradient exploited by EC 7.1.2.2 for ATP generation. A representative reaction for EC 7.1.2.2 in the synthesis mode is: where is typically 3–4 protons, highlighting the stoichiometry that links proton flow to phosphorylation potential. Similarly, for EC 7.1.1.2, the reaction is: with Q denoting ubiquinone, underscoring the redox-driven proton pumping.[21]EC 7.2: Inorganic Cation Translocation
EC 7.2 enzymes catalyze the translocation of inorganic cations, such as Na^+, K^+, Ca^{2+}, and their chelates, across biological membranes from one side (side 1) to the other (side 2), often powered by energy sources like nucleoside triphosphate hydrolysis or redox reactions. These translocases play essential roles in maintaining cellular ion homeostasis, including osmotic balance, by actively transporting cations against their electrochemical gradients. Subclasses include EC 7.2.1 (linked to oxidoreductase reactions), EC 7.2.2 (linked to nucleoside triphosphate hydrolysis), EC 7.2.3 (linked to diphosphate hydrolysis), and EC 7.2.4 (linked to decarboxylation), encompassing 31 distinct enzyme entries as of May 2023.[18][22] A prominent subclass is EC 7.2.2, where translocation is coupled to the hydrolysis of ATP or other nucleoside triphosphates, enabling primary active transport of cations. For instance, the plasma membrane Ca^{2+} ATPase (PMCA, EC 7.2.2.10) extrudes Ca^{2+} from the cytosol to the extracellular space, helping to keep intracellular Ca^{2+} levels low for proper signaling and preventing osmotic perturbations due to ion imbalances. The reaction catalyzed by PMCA is: This P-type ATPase undergoes autophosphorylation during its transport cycle, moving 1 Ca^{2+} ion per ATP hydrolyzed.[23][24] Another key example is the Na^+/K^+-exchanging ATPase (EC 7.2.2.13), which maintains the Na^+ and K^+ gradients essential for membrane potential and osmotic equilibrium in animal cells. This enzyme exchanges three Na^+ ions out for two K^+ ions in per ATP molecule hydrolyzed, counteracting passive ion leaks and supporting cell volume regulation. The reaction is: The gastric H^+/K^+-exchanging ATPase (EC 7.2.2.19), a related P-type pump, facilitates acid secretion in parietal cells by exchanging H^+ for K^+, further illustrating the diversity of cation-specific transport in EC 7.2. These mechanisms are critical for physiological processes like nerve impulse transmission and muscle contraction, where disruptions in cation gradients can lead to cellular swelling or shrinkage. Some EC 7.2 enzymes, such as certain Na^+-transporting variants, may couple cation movement with proton gradients established by EC 7.1 hydron translocases.[25][26]EC 7.3: Inorganic Anion Translocation
EC 7.3 comprises translocases that catalyze the movement of inorganic anions and their chelates across biological membranes, typically from one side of the membrane to the other.[18] These enzymes are primarily ATP-binding cassette (ABC)-type transporters that couple anion translocation to the hydrolysis of nucleoside triphosphates, such as ATP, enabling active transport against concentration gradients.[27] The subclass focuses on inorganic species including phosphate (PO₄³⁻), sulfate (SO₄²⁻), nitrate (NO₃⁻), molybdate (MoO₄²⁻), and arsenite (AsO₃³⁻), distinguishing it from transporters of organic anions or cations classified elsewhere.[28] Currently, EC 7.3 includes seven distinct enzymes, all assigned to the sub-subclass EC 7.3.2, which links translocation to nucleoside triphosphate hydrolysis.[27] These encompass ABC-type transporters for phosphate (EC 7.3.2.1), phosphonate (EC 7.3.2.2), sulfate (EC 7.3.2.3), nitrate (EC 7.3.2.4), molybdate (EC 7.3.2.5), and tungstate (EC 7.3.2.6), as well as an arsenite-transporting ATPase (EC 7.3.2.7).[18] Functionally, these translocases play critical roles in microbial nutrient acquisition, such as importing phosphate essential for nucleic acid synthesis and energy metabolism, or sulfate for cysteine and methionine biosynthesis.[29] Additionally, they contribute to waste export, exemplified by the efflux of toxic arsenite to maintain cellular homeostasis.[30] A representative example is the ABC-type sulfate transporter (EC 7.3.2.3), which facilitates high-affinity uptake of sulfate and thiosulfate in bacteria like Escherichia coli by interacting with periplasmic binding proteins.[31] The catalyzed reaction is: This process powers sulfate assimilation pathways, underscoring the enzyme's importance in sulfur metabolism.[32] Similarly, the ABC-type phosphate transporter (EC 7.3.2.1) supports phosphorus homeostasis through an analogous ATP-driven mechanism, without involving proton symport.[29]EC 7.4: Amino Acid and Peptide Translocation
EC 7.4 encompasses enzymes that catalyze the translocation of amino acids and short peptides across biological membranes, primarily in prokaryotes, mitochondria, and chloroplasts. These translocases are essential for the directed movement of nitrogen-containing organic molecules, enabling cells to acquire vital building blocks for protein synthesis and metabolism. The subclass EC 7.4.2 specifically links this translocation to the hydrolysis of nucleoside triphosphates, most commonly ATP, providing the energy required to drive transport against concentration gradients.[33][18] Currently, the International Union of Biochemistry and Molecular Biology (IUBMB) recognizes 14 distinct enzymes within EC 7.4, all falling under the ATP-dependent subclass EC 7.4.2. These include prominent examples such as EC 7.4.2.1 (ABC-type polar amino acid transporter), which facilitates the import of polar amino acids like glutamine, histidine, and arginine in bacteria, and EC 7.4.2.6 (ABC-type oligopeptide transporter), involved in the uptake of short peptides up to five residues long. A representative reaction for these enzymes is:amino acid [side 1] + ATP + H₂O → amino acid [side 2] + ADP + phosphate,
where "side 1" and "side 2" denote the opposing faces of the membrane. This ATP-driven mechanism typically involves a multi-subunit ABC transporter complex, comprising transmembrane domains for substrate recognition and translocation, and nucleotide-binding domains for ATP hydrolysis.[18][34][35] These translocases exhibit high specificity for charged or polar amino acid residues and short peptides, often interacting with periplasmic substrate-binding proteins in Gram-negative bacteria to selectively capture scarce nutrients from the extracellular environment. This specificity underscores their critical role in nutrient scavenging, particularly in nutrient-poor habitats, where they support bacterial growth, virulence, and adaptation by enabling the efficient uptake of amino acids and peptides derived from host proteins or environmental sources. For instance, in pathogenic bacteria like Escherichia coli and Staphylococcus aureus, ABC-type amino acid transporters under EC 7.4 contribute to pathogenesis by sustaining metabolism during infection.[36][37]

