Recent from talks
Nothing was collected or created yet.
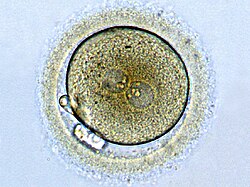
Zygote
View on Wikipedia| Zygote | |
|---|---|
 Zygote formation: egg cell after fertilization with a sperm. The male and female pronuclei are converging, but the genetic material is not yet united. | |
| Details | |
| Days | 0 |
| Precursor | Gametes |
| Gives rise to | Blastomeres |
| Identifiers | |
| MeSH | D015053 |
| TE | E2.0.1.2.0.0.9 |
| FMA | 72395 |
| Anatomical terminology | |
| Part of a series on |
| Human growth and development |
|---|
 |
| Stages |
| Biological milestones |
| Development and psychology |
A zygote (/ˈzaɪˌɡoʊt/ ⓘ; from Ancient Greek ζυγωτός (zygōtós) 'joined, yoked', from ζυγοῦν (zygoun) 'to join, to yoke')[1] is a eukaryotic cell formed by a fertilization event between two gametes.
The zygote's genome is a combination of the DNA in each gamete, and contains all of the genetic information of a new individual organism.
The sexual fusion of haploid cells is called karyogamy, the result of which is the formation of a diploid cell called the zygote or zygospore.
History
[edit]German zoologists Oscar and Richard Hertwig made some of the first discoveries on animal zygote formation in the late 19th century.[citation needed]
In multicellular organisms
[edit]The zygote is the earliest developmental stage. In humans and most other anisogamous organisms, a zygote is formed when an egg cell and sperm cell come together to create a new unique organism.[2]
The formation of a totipotent zygote with the potential to produce a whole organism depends on epigenetic reprogramming. DNA demethylation of the paternal genome in the zygote appears to be an important part of epigenetic reprogramming.[3] In the paternal genome of the mouse, demethylation of DNA, particularly at sites of methylated cytosines, is likely a key process in establishing totipotency. Demethylation involves the processes of base excision repair and possibly other DNA-repair–based mechanisms.[3]
Humans
[edit]
In human fertilization, a released ovum (a haploid secondary oocyte with replicate chromosome copies) and a haploid sperm cell (male gamete) combine to form a single diploid cell called the zygote. Once the single sperm fuses with the oocyte, the latter completes the division of the second meiosis forming a haploid daughter with only 23 chromosomes, almost all of the cytoplasm, and the male pronucleus. The other product of meiosis is the second polar body with only chromosomes but no ability to replicate or survive. In the fertilized daughter, DNA is then replicated in the two separate pronuclei derived from the sperm and ovum, making the zygote's chromosome number temporarily 4n diploid. After approximately 30 hours from the time of fertilization, a fusion of the pronuclei and immediate mitotic division produce two 2n diploid daughter cells called blastomeres.[4] Between the stages of fertilization and implantation, the developing embryo is sometimes termed as a preimplantation-conceptus. This stage has also been referred to as the pre-embryo in legal discourses including relevance to the use of embryonic stem cells.[5] In the US the National Institutes of Health has determined that the traditional classification of pre-implantation embryo is still correct.[6]
After fertilization, the conceptus travels down the fallopian tube towards the uterus while continuing to divide[7] without actually increasing in size, in a process called cleavage.[8] After four divisions, the conceptus consists of 16 blastomeres, and it is known as the morula.[9] Through the processes of compaction, cell division, and blastulation, the conceptus takes the form of the blastocyst by the fifth day of development, just as it approaches the site of implantation.[10] When the blastocyst hatches from the zona pellucida, it can implant in the endometrial lining of the uterus and begin the gastrulation stage of embryonic development.[citation needed]
The human zygote has been genetically edited in experiments designed to cure inherited diseases.[11]
Fungi
[edit]In fungi, this cell may then enter meiosis or mitosis depending on the life cycle of the species.[citation needed]
Plants
[edit]In plants, the zygote may be polyploid if fertilization occurs between meiotically unreduced gametes.[12]
In land plants, the zygote is formed within a chamber called the archegonium. In seedless plants, the archegonium is usually flask-shaped, with a long hollow neck through which the sperm cell enters. As the zygote divides and grows, it does so inside the archegonium.[citation needed]
In single-celled organisms
[edit]The zygote can divide asexually by mitosis to produce identical offspring.[citation needed]
A Chlamydomonas zygote contains chloroplast DNA (cpDNA) from both parents; such cells are generally rare, since normally cpDNA is inherited uniparentally from the mt+ mating type parent. These rare biparental zygotes allowed mapping of chloroplast genes by recombination.[citation needed]
See also
[edit]References
[edit]- ^ "English etymology of zygote". etymonline.com. Archived from the original on 2017-03-30.
- ^ Khan, Yusuf S.; Ackerman, Kristin M. (2025), "Embryology, Week 1", StatPearls, Treasure Island (FL): StatPearls Publishing, PMID 32119449, retrieved 2025-07-02
- ^ a b Ladstätter S, Tachibana-Konwalski K (December 2016). "A Surveillance Mechanism Ensures Repair of DNA Lesions during Zygotic Reprogramming". Cell. 167 (7): 1774–1787.e13. doi:10.1016/j.cell.2016.11.009. PMC 5161750. PMID 27916276.
- ^ Blastomere Encyclopædia Britannica Archived 2013-09-28 at the Wayback Machine. Encyclopædia Britannica Online. Encyclopædia Britannica Inc., 2012. Web. 06 Feb. 2012.
- ^ Condic, Maureen L. (14 April 2014). "Totipotency: What It Is And What It Is Not". Stem Cells and Development. 23 (8): 796–812. doi:10.1089/scd.2013.0364. PMC 3991987. PMID 24368070.
- ^ "Report of the Human Embryo Research Panel" (PDF). Archived from the original (PDF) on 2009-01-30. Retrieved 2009-02-17.
- ^ O'Reilly, Deirdre. "Fetal development ". MedlinePlus Medical Encyclopedia (2007-10-19). Retrieved 2009-02-15.
- ^ Klossner, N. Jayne and Hatfield, Nancy. Introductory Maternity & Pediatric Nursing, p. 107 (Lippincott Williams & Wilkins, 2006).
- ^ Neas, John F. "Human Development" Archived July 22, 2011, at the Wayback Machine. Embryology Atlas
- ^ Blackburn, Susan. Maternal, Fetal, & Neonatal Physiology, p. 80 (Elsevier Health Sciences 2007).
- ^ "Editing human germline cells sparks ethics debate". May 6, 2015. Archived from the original on May 18, 2015. Retrieved May 17, 2020.
- ^ Brownfield, Lynette; Köhler, Claudia (2011-03-01). "Unreduced gamete formation in plants: mechanisms and prospects". Journal of Experimental Botany. 62 (5): 1659–1668. doi:10.1093/jxb/erq371. hdl:20.500.11850/31348. ISSN 0022-0957.
Zygote
View on GrokipediaEtymology and History
Terminology and Discovery
The term zygote originates from the Ancient Greek zygōtós (ζυγωτός), meaning "yoked" or "joined," alluding to the conjugation of gametes in reproduction.[9] [10] This etymological root reflects the process of fusion between male and female reproductive cells, a concept formalized in late 19th-century biology. The word entered scientific usage through German botanist Eduard Strasburger, who coined Zygote in 1878 to describe a fused spore in fungi and algae, later extending it to the fertilized ovum in animals.[9] Early identification of the zygote's components relied on microscopic advancements. In 1677, Dutch microscopist Antonie van Leeuwenhoek first observed and described spermatozoa in human semen, revealing motile "animalcules" as potential contributors to generation.[11] [12] This discovery challenged preformationist theories but did not yet link sperm to egg fusion. Fifty years later, in 1827, Karl Ernst von Baer identified the mammalian ovum by examining ovarian follicles in dogs, confirming its presence across mammals including humans and resolving debates over whether such eggs existed in viviparous species.[13] [14] The zygote as a distinct entity—the single cell resulting from gamete union—emerged with 1870s research on fertilization mechanics. German embryologist Oscar Hertwig, studying sea urchin eggs, observed in 1876 that a single spermatozoon penetrates the ovum, leading to nuclear fusion and the formation of a diploid nucleus, which he identified as the critical initial step in development.[15] [16] This work, disseminated in embryology literature by the 1880s, shifted understanding from mere mixing of substances to precise cellular amalgamation, establishing the zygote's foundational role without invoking later genetic details.[15]Key Milestones in Research
In 1876, Oscar Hertwig's microscopic observations of sea urchin (Echinus microtuberculatus) eggs demonstrated that fertilization entails the fusion of sperm and egg pronuclei, forming a single diploid nucleus that initiates embryonic development and resolves prior debates on parthenogenesis versus syngamy.[17] This empirical breakthrough established the zygote as the product of nuclear amalgamation, providing direct evidence against theories of sperm as mere activators without genetic contribution.[18] In 1924, Hans Spemann and Hilde Mangold's transplantation experiments on newt embryos identified the "organizer" in the dorsal lip of the blastopore, showing how cells derived from the zygote emit inductive signals that direct axis formation and tissue differentiation in neighboring regions.[19] Their work, earning Spemann the 1935 Nobel Prize, highlighted the zygote's foundational role in generating regulatory interactions essential for ordered embryogenesis, shifting focus from mosaic to regulative development models.[20] The 1953 discovery of DNA's double-helix structure by James Watson and Francis Crick enabled molecular dissection of the zygote's genome, revealing how complementary base pairing ensures stable inheritance of the fused haploid sets during replication and transcription.[21] This framework spurred subsequent investigations into zygotic genome activation, confirming that the diploid complement dictates developmental potential independent of cytoplasmic factors alone.[22] On July 25, 1978, the birth of Louise Brown, the first human conceived via in vitro fertilization by Robert Edwards, Patrick Steptoe, and Jean Purdy, empirically verified the zygote's viability and full developmental competence when formed and initially cultured ex vivo before transfer.30261-9/fulltext) This achievement, involving over 100 failed attempts, underscored the zygote's intrinsic robustness against artificial conditions, paving the way for controlled studies of early cleavage without maternal physiological dependencies.[23]Definition and Characteristics
Biological Definition
A zygote is the single diploid cell formed immediately following the syngamy, or fusion, of two haploid gametes—a spermatozoon and an oocyte—during fertilization in sexually reproducing organisms.[8] This fusion restores the diploid chromosome number (2n), combining the haploid genomes (n) from each parent to create the complete genetic complement necessary for embryonic development.[24] The zygote represents the foundational unit of the new organism, distinct from its parental gametes by virtue of its restored ploidy and initiation of autonomous cellular processes.[25] In contrast to gametes, which are specialized haploid cells produced via meiosis and incapable of independent development beyond fusion, the zygote exhibits totipotency—the capacity to differentiate into every cell type of the organism, including extraembryonic tissues.[26] Gametes serve exclusively as vehicles for genetic transmission, lacking the machinery for sustained division or morphogenesis until syngamy occurs.[27] Empirical markers of the zygote include its diploid karyotype, verifiable through cytogenetic analysis, and a unique genotypic profile resulting from allelic combinations and recombination events in gametogenesis, setting it apart from either parent's genome.[24] The zygote's formation marks the transition from gametic to embryonic phases, with its cellular integrity confirmed by the presence of pronuclei fusion and subsequent decondensation of chromatin, observable via microscopy in model organisms.[8] This initial cell encapsulates the organism's full developmental potential, grounded in its genomic completeness rather than extrinsic maternal provisions alone.[25]Cellular and Genetic Properties
The zygote constitutes a single diploid eukaryotic cell, featuring a voluminous cytoplasm predominantly derived from the oocyte, which endows it with substantial reserves of mitochondria, ribosomes, endoplasmic reticulum, and cortical granules essential for metabolic autonomy prior to embryonic genome activation.[25] This cytoplasmic abundance, often exceeding 100 micrometers in diameter in mammalian species, supports the cell's initial synthetic demands and protects the nuclear material during early mitotic preparations.[28] Genetically, the zygote harbors a complete diploid set of chromosomes—46 in humans, comprising 23 pairs—integrating the haploid contributions from each gamete without immediate recombination beyond fertilization-induced events.[29] Epigenetic modifications, including DNA methylation patterns and histone variants, are asymmetrically inherited: maternal imprints from the oocyte silence or activate specific loci via differentially methylated regions (DMRs), while paternal marks from sperm often favor expression of growth-promoting genes, thereby establishing parent-of-origin-specific gene regulation that persists through development.[30] These imprints, verifiable through bisulfite sequencing in isolated zygotes, number approximately 100-200 imprinted genes in mammals, influencing phenomena like fetal growth without altering the DNA sequence itself.[31] The zygote exemplifies totipotency, defined experimentally as the capacity to generate all embryonic and extraembryonic lineages, evidenced by its division into a blastocyst capable of implanting and forming a viable organism in vivo, or by microsurgical isolation of early blastomeres yielding chimeric progeny in model organisms.[32] This potential, rooted in unrestricted access to the full genomic repertoire and minimal lineage commitment, contrasts with later pluripotent states and is molecularly underpinned by transient expression of factors like Zscan4 and high mobility group proteins that maintain chromatin openness.[26] In humans, totipotency is inferred from preimplantation embryo transfers succeeding in full-term births, with failure rates attributable to extrinsic factors rather than intrinsic limitations.[33]Formation Process
Fertilization Mechanics
Fertilization initiates when a sperm cell contacts the zona pellucida surrounding the oocyte, triggering species-specific binding that induces the acrosome reaction in the sperm head.[24] This reaction releases hydrolytic enzymes, such as acrosin and hyaluronidase, from the acrosomal vesicle, enabling the sperm to digest and penetrate the zona pellucida matrix.[24] The process relies on actin polymerization in the sperm to propel it through the zona.[24] Following zona penetration, the sperm's plasma membrane fuses with the oocyte's plasma membrane, typically at the equatorial region of the sperm head.[34] This fusion delivers sperm contents, including a soluble factor like phospholipase C zeta (PLCζ), into the oocyte cytoplasm.[35] Membrane fusion is mediated by proteins such as Izumo1 on sperm and JUNO on the oocyte, forming a transient complex essential for gamete union.[36] Sperm-oocyte fusion promptly triggers intracellular calcium (Ca²⁺) oscillations in the oocyte, rising more than tenfold and persisting for minutes to hours.[37] These oscillations, driven by inositol 1,4,5-trisphosphate (IP₃) production from PLCζ activity, signal oocyte activation and induce the cortical reaction, where cortical granules exocytose contents that modify the zona pellucida glycoproteins, hardening it and destroying sperm receptors to block polyspermy.[34][35] The calcium signals also resume oocyte meiosis, extruding the second polar body and completing the second meiotic division, while promoting decondensation of the highly compacted sperm nucleus through disassembly of protamine-stabilized chromatin.[38][39] This decondensation involves histone replacement and nuclear envelope reformation, marking the transition to zygotic formation without yet addressing genetic reprogramming.[40]Post-Fusion Events
Immediately following the fusion of sperm and oocyte plasma membranes, the sperm nucleus decondenses as protamines are replaced by histones, forming the male pronucleus, while the oocyte completes meiosis II, extruding the second polar body and forming the female pronucleus.[41] This decondensation is facilitated by oocyte factors such as glutathione and nucleoplasmin, enabling chromatin remodeling within minutes to hours post-fusion.[42] The sperm contributes the functional centrosome, which nucleates microtubules to form the sperm aster; this structure drives pronuclear migration by interacting with the oocyte cortex and centering the pronuclei.[43] Microscopy studies in mammalian zygotes, including humans, confirm the sperm-derived centrioles reconstitute the zygotic centrosome, essential for organizing the microtubule network that apposes the pronuclei prior to syngamy.[44] In human zygotes, this centrosomal activity ensures proper setup for the first mitotic spindle, distinct from acentrosomal mechanisms in oocytes.[45] DNA synthesis initiates asynchronously in the pronuclei, with paternal genome replication beginning earlier than maternal due to sperm chromatin protamine removal; in mouse zygotes, this occurs 5-6 hours post-fertilization, preceding pronuclear fusion.[46] Human studies indicate S-phase entry around 10-14 hours after insemination, during or shortly after pronuclear migration, marking the transition to zygotic genome activation preparation.[47] Errors in these post-fusion processes, such as defective pronuclear congression or centrosome malfunction, contribute to aneuploidy; in human cleavage-stage embryos, parental genome unification failures result in 50-70% aneuploidy rates, often from chromosome misalignment during the first mitosis.[48] Empirical data from IVF embryos show the initial mitotic division is particularly error-prone, with spindle assembly defects amplifying meiotic errors into zygotic mosaicism.[49] These rates underscore the vulnerability of early zygotic microtubule dynamics to perturbations, independent of gamete quality.[50]Developmental Role
Zygotic Activation
Zygotic genome activation (ZGA) constitutes the onset of embryonic transcription from the zygotic genome, enabling the shift from reliance on maternally inherited mRNAs and proteins to self-directed gene expression during the maternal-to-zygotic transition (MZT).[51] This process ensures the embryo gains transcriptional independence, with zygotic transcripts replacing degraded maternal factors to drive subsequent development.[52] Empirical studies using RNA sequencing have quantified this handover, revealing coordinated downregulation of thousands of maternal mRNAs alongside upregulation of zygotic genes essential for cell cycle progression and metabolism.[53] Mechanistically, ZGA involves the targeted degradation of maternal mRNAs, mediated by pathways such as deadenylation, decapping, and exonucleolytic cleavage, which clear oocyte-stored transcripts to prevent interference with embryonic programs.[54] RNA sequencing analyses in model systems have empirically confirmed this degradation's timing and scope, showing that maternal transcripts decline sharply as zygotic transcription ramps up, with specific factors like YTHDF2 promoting selective mRNA destabilization post-fertilization.[55] Concurrently, chromatin remodeling and RNA polymerase II recruitment facilitate zygotic promoter activation, marking a causal pivot to embryonic control independent of maternal cues.[56] In mammals, ZGA exhibits species-specific timing reflective of cleavage rates and developmental tempo: minor activation emerges at the late 1-cell stage in mice, escalating to major ZGA by the 2- to 4-cell stages, whereas in humans, robust transcription initiates predominantly at the 8-cell stage.[57][58] Across vertebrates, variations persist—earlier in teleost fish and amphibians relative to elapsed time, but universally tied to cell size reductions rather than division counts—yet all underscore a conserved causal necessity for metabolic autonomy from maternal provisioning.[52][59] Disruptions in this activation, as evidenced by transcription inhibitors, halt development, affirming its empirical role in viability.[60]Cleavage and Early Division
Following fertilization, the zygote undergoes cleavage, a series of rapid mitotic divisions that partition the cytoplasm into progressively smaller blastomeres without a net increase in overall embryo size or mass.[61] In mammals, including humans, this process features holoblastic cleavage, where the entire zygote divides completely due to the minimal yolk content in the egg, resulting in equal or near-equal blastomere sizes initially. The first cleavage typically occurs approximately 24 hours post-fertilization, yielding a 2-cell stage, followed by subsequent divisions to 4-cell (around 40 hours), 8-cell (around 72 hours), and 16-cell stages.[5] Early cleavage cycles are abbreviated, oscillating primarily between S (DNA synthesis) and M (mitosis) phases, with the absence of a G1 gap phase enabling accelerated division rates that shorten from ~20-24 hours initially to as little as 10-12 hours by the 8-cell stage.[62] This rapid progression culminates in the morula stage by days 3-4 post-fertilization, a compact ball of 16-32 blastomeres where cell volume decreases as divisions continue without interstitial growth.[63] Compaction begins around the 8-cell stage in human embryos, driven by cell-cell adhesion via E-cadherin and cytoskeletal changes, transforming the loosely arranged blastomeres into a tighter spherical mass that facilitates further differentiation cues.[64] By day 5, the morula transitions to the blastocyst, comprising 50-150 cells with fluid accumulation forming a blastocoel cavity and differentiation into an outer trophoblast layer and an inner cell mass (ICM) that will contribute to the fetus proper; this stage is routinely observed in in vitro fertilization (IVF) protocols as a key viability marker.[5] Throughout cleavage, the embryo's total cytoplasmic content remains conserved, emphasizing equitable partitioning over biomass accumulation to establish the foundational multicellular architecture.[65]Zygote Across Organisms
In Multicellular Animals
In multicellular animals, the zygote represents the initial totipotent cell stage following fertilization, capable of differentiating into all cell types of the embryo proper as well as extraembryonic structures such as trophoblast or membranes.[26] This totipotency persists briefly into early blastomeres in species like mice, where single cells from the two- or four-cell stage can develop into viable offspring when supported appropriately.[33] Unlike plant zygotes, which arise from double fertilization and contribute to both embryo and nutritive endosperm tissues within a protective ovule, animal zygotes undergo direct cleavage without such endosperm formation, relying instead on maternal yolk provisions or post-fertilization nutrient uptake.[66] Cleavage patterns in animal zygotes vary primarily due to yolk amount and distribution, which mechanically constrains cell division. In eggs with little to moderate yolk (isolecithal or mesolecithal, as in many invertebrates, mammals, and amphibians), holoblastic cleavage divides the entire zygote into smaller blastomeres, often in radial or spiral arrangements that establish early polarity.[61] Conversely, in telolecithal eggs with abundant vegetal yolk, such as those of birds, reptiles, and some fish, meroblastic cleavage is restricted to the animal pole, forming a blastodisc atop the undivided yolk mass to accommodate the large nutrient reserve.[67] These yolk-driven differences contrast with fungal zygotes, which typically undergo immediate meiosis or dikaryotic growth rather than iterative mitotic cleavages leading to multicellular embryos.[66] Despite cleavage variability, animal zygotes share conserved genetic mechanisms for initiating development, including maternal-to-zygotic transition where zygotic transcription activates core patterning genes. Hox gene clusters, highly preserved across metazoans, begin influencing anterior-posterior axis formation shortly after zygotic genome activation, underpinning the shared bilaterian body plans observed empirically in diverse phyla from cnidarians to chordates.[68] This conservation reflects ancient evolutionary origins, with Hox-like genes detectable in pre-bilaterian animals, enabling causal coordination of segment identity independent of yolk-imposed morphological constraints.[69]In Humans Specifically
The human zygote arises from the fusion of a spermatozoon with a secondary oocyte, typically within the ampulla of the fallopian tube, completing the fertilization process approximately 24 hours after ovulation and insemination.[70] This diploid cell contains 46 chromosomes, comprising 23 pairs derived from the maternal and paternal genomes.[3] Empirical data from in vitro fertilization (IVF) procedures corroborate this timeline, as pronuclei fusion and syngamy are observed within 18-24 hours post-insemination in cultured oocytes.[70] Aneuploidy poses a significant challenge in human zygotes, with IVF-derived studies reporting rates up to 50% in early embryos due to meiotic errors in gametes or mitotic segregation failures shortly after fertilization.[71] This chromosomal instability contributes to pre-implantation developmental arrest, with approximately 20-40% of zygotes failing to progress to the blastocyst stage in natural cycles, as inferred from high attrition rates observed in assisted reproduction.[71] Preimplantation genetic testing data further indicate that aneuploidy prevalence increases with maternal age, exceeding 70% in women over 35 years.[72] Zygotic genome activation (ZGA) in human embryos initiates with minor transcriptional bursts at the one-cell stage but escalates to major activation between the 4- and 8-cell stages, marking the transition from maternal to embryonic genetic control.[73] Single-cell RNA sequencing from IVF embryos confirms this pattern, revealing upregulation of thousands of zygotic genes by the 8-cell stage.[58] Following ZGA, the zygote undergoes asynchronous cleavage divisions, forming a compact morula by day 3 post-fertilization and a fluid-filled blastocyst by day 5-6.[74] The blastocyst subsequently hatches from the zona pellucida around day 6, a process essential for uterine implantation, as documented in time-lapse imaging of IVF embryos where hatching correlates with trophectoderm expansion and zona thinning.[75] This event enables direct endometrial contact, with successful hatching observed in 20-30% of cultured blastocysts under standard IVF conditions.[74]In Plants and Fungi
In angiosperms, double fertilization occurs within the embryo sac of the ovule, where one sperm cell fuses with the haploid egg cell to form a diploid zygote destined to develop into the embryo, while a second sperm cell fuses with the diploid central cell to produce triploid endosperm that nourishes the developing seed.[76] This process, unique to flowering plants, ensures coordinated development of embryonic tissues.[77] Following fertilization, the plant zygote elongates and undergoes a characteristic asymmetric transverse division, yielding a smaller apical daughter cell that proliferates to form the embryo proper and a larger basal daughter cell that differentiates into the suspensor—a transient structure that anchors the embryo and facilitates nutrient transfer from maternal tissues.[78] This polarity establishes the apical-basal axis essential for subsequent embryogenesis, diverging from symmetric divisions in many animal zygotes by prioritizing extra-embryonic support early on.[79] In fungi, zygote formation varies by taxon but exemplifies brief diploidy in haplontic cycles, as seen in ascomycete yeasts like Saccharomyces cerevisiae, where haploid cells of complementary mating types (a and α) fuse via shmoo formation to create a diploid zygote.[80] This zygote, the sole diploid entity in the cycle, rapidly initiates meiosis under nutrient stress, producing a tetrad of haploid ascospores that germinate into mitotic haploid clones, thus minimizing the diploid phase compared to the prolonged embryonic development in plants.[81] Such immediate meiotic commitment reflects adaptive divergence, prioritizing genetic recombination over diploid growth in these unicellular-dominant fungi.[82]In Unicellular Organisms
In ciliates such as Paramecium, conjugation serves as the primary sexual process, involving the temporary pairing of two cells of compatible mating types. Each cell contributes a migratory haploid micronucleus that is exchanged and fuses with a stationary haploid micronucleus in the partner cell, yielding a diploid synkaryon, or zygote nucleus.[83] This occurs after prezygotic mitotic divisions of the micronuclei, with the resulting zygote nucleus undergoing three postzygotic divisions: the products differentiate into new macronuclei (via amitosis) and micronuclei, while the original macronuclei degenerate.[84] The process restores nuclear dimorphism without forming a persistent diploid cell lineage, differing from metazoan zygotes by emphasizing nuclear reorganization over cytoplasmic fusion.[83] In unicellular green algae like Chlamydomonas reinhardtii, zygote formation arises from the fusion of isogametes or anisogametes of opposite mating types (mt+ and mt-), producing a quadriflagellate diploid zygote that soon encysts into a thick-walled zygospore.[85] This establishes temporary diploidy, during which the zygote undergoes extensive remodeling, including selective degradation of parental chloroplast DNA and activation of zygote-specific genes.[86] Meiosis follows germination, yielding haploid spores that develop into vegetative cells, as evidenced by genetic complementation assays using auxotrophic markers that confirm biparental inheritance and fusion events.[85] Such mechanisms parallel gametic fusion in multicellular eukaryotes but terminate in haploid progeny without embryonic cleavage or tissue formation.[87]Research Applications and Advances
In Reproductive Technologies
In in vitro fertilization (IVF), mature oocytes retrieved from the ovaries are combined with sperm in a laboratory dish, leading to fertilization and the formation of zygotes within 24 hours. These zygotes are then cultured under controlled conditions to monitor early cleavage divisions, typically reaching the 4- to 8-cell stage before potential transfer or further development to the blastocyst stage on day 5. Zygote intrafallopian transfer (ZIFT), a variant, involves placing the zygote directly into the fallopian tube to mimic natural transport, though it is less commonly performed today due to advancements in uterine transfer techniques.[88][89] Intracytoplasmic sperm injection (ICSI), used primarily for male infertility factors such as low sperm count or motility, bypasses natural barriers by injecting a single spermatozoon directly into the oocyte cytoplasm, resulting in zygote formation with fertilization rates of 50-80%. This technique achieves higher fertilization success compared to conventional IVF insemination, particularly in cases of severe oligospermia, and the resulting zygotes exhibit kinetics that may accelerate early development. Post-ICSI zygotes are cultured similarly to standard IVF zygotes, with pronuclear verification confirming successful fertilization.[90][91] To enhance implantation success and reduce aneuploidy-related losses, preimplantation genetic testing for aneuploidy (PGT-A, formerly PGS) screens embryos derived from cultured zygotes for chromosomal abnormalities via biopsy, typically at the blastocyst stage, allowing selection of euploid embryos for transfer. Empirical live birth rates per IVF cycle, incorporating these technologies, range from 30-50% for women under 35 years old, depending on clinic protocols and patient age, with single embryo transfers yielding around 25% per fresh transfer in recent UK data and higher cumulative rates over multiple cycles. Success varies, with clinics reporting up to 58% per retrieval for optimal candidates, reflecting improvements in zygote handling and genetic selection.[92][93][94]Recent Models and Synthetic Approaches
In 2023, researchers generated synthetic human embryo models from extended pluripotent stem cells (EPSCs), which self-organize into structures encompassing embryonic and extraembryonic lineages without gamete fusion or fertilization, mimicking early post-zygotic development up to day 14 with features including yolk sac formation and primitive streak initiation.[95] These models demonstrate biological fidelity in gene expression profiles and morphological organization comparable to natural embryos, enabling analysis of implantation and gastrulation mechanisms.[96] Subsequent refinements in 2025 have integrated hypoblast-like tissues, enhancing recapitulation of amniotic cavity development while relying on transgene-free protocols from embryonic stem cells.[97][98] Stem cell-derived blastoids, aggregating naive pluripotent stem cells to form blastocyst-like entities, bypass zygote formation by directly specifying trophectoderm, epiblast, and primitive endoderm, achieving transcriptional and structural similarity to natural blastocysts in over 80% of lineage-specific markers.[99] Advances post-2023 include scalable production protocols yielding thousands of human blastoids for high-throughput studies of peri-implantation events, with 2025 reports confirming their capacity for post-implantation progression under 3D matrix culture, including epiblast cavitation and trophoblast invasion.00285-0)00288-6) These models have validated ~90% concordance in DNA methylation dynamics with in vivo counterparts, prioritizing empirical metrics over ethical proxies for developmental fidelity.[100] CRISPR-Cas9 applications in synthetic and animal zygotes have progressed with 2024-2025 optimizations reducing off-target effects to below 1% via enhanced guide RNA designs and base editing variants, tested in porcine models for monogenic trait correction relevant to disease modeling.[101] In mouse blastoid systems, multiplex editing has enabled dissection of zygotic genome activation pathways, with efficiency rates exceeding 70% for homozygous modifications, informing causal roles in early lineage commitment without human embryo involvement.[102] These approaches emphasize verifiable editing precision over speculative therapeutic translation, constrained by regulatory moratoriums on heritable human edits.00111-9)Biological Status and Debates
Scientific Consensus on Ontogeny
The zygote forms at fertilization when the sperm and egg unite, creating a genetically unique diploid cell that initiates the ontogeny of a new multicellular organism, distinct from the gametes that produced it. Embryological texts consistently describe this event as the commencement of development, with the zygote possessing a complete genome directing subsequent cellular divisions and differentiation without requiring external genetic input.[103][104] This genetic individuality—combining half the chromosomes from each parent—establishes the zygote as the foundational entity of the organism's life cycle, verifiable through DNA sequencing that confirms its novel allelic combinations absent in either gamete.[70] Empirical surveys of biologists reinforce this view: a 2023 analysis of responses from 5,577 experts across 1,058 institutions worldwide found 96% agreement that fertilization marks the beginning of a human organism's life, based on criteria like genomic activation and developmental continuity.[105][106] Such consensus derives from observable biological markers, including the zygote's immediate metabolic autonomy and programmed cleavage stages, rather than later milestones like implantation or organogenesis. The zygote's totipotency further evidences its self-directing nature, as it can generate all embryonic and extraembryonic lineages, sustaining uninterrupted development toward maturity through intrinsic regulatory mechanisms.[32][107] Experimental perturbations, such as isolating blastomeres in mammalian models, demonstrate this potential persists briefly post-cleavage, confirming the zygote's embedded program as causally sufficient for ontogenetic progression.[33] This continuity—evident in the absence of type changes or external organismal fusion—aligns with causal chains observed in vivo, from zygotic genome activation to adult morphology.[108]Viewpoints on Organismal Status
The biological perspective, rooted in embryology, holds that the zygote constitutes a distinct human organism from the moment of fertilization, when the sperm and egg fuse to form a unique diploid genome directing self-organized development.[109][110] This view counters characterizations of the zygote as a mere "clump of cells" by emphasizing its totipotency, genomic individuality separate from parental DNA, and initiation of continuous ontogenetic processes toward a mature human form, without external imposition of organismal identity.[4][7] Proponents, including those aligned with pro-life positions, argue this marks the onset of human life as an individuated member of Homo sapiens, supported by standard embryological texts and surveys indicating biologist consensus on fertilization as the starting point.[105][109] Opposing viewpoints often decouple biological organismal status from moral or legal personhood, proposing delayed thresholds such as implantation (around 5-10 days post-fertilization), viability (approximately 24 weeks gestation, when survival outside the womb becomes possible with medical aid), or sentience (linked to neural development around the same period).[111][112] Advocates of these criteria, frequently from philosophical or ethical frameworks, contend that personhood requires capacities like consciousness, viability independent of maternal support, or relational viability, dismissing the zygote's status due to its dependence and lack of such traits.[113][114] Religious supplements, such as certain interpretations emphasizing ensoulment at quickening or birth, or secular philosophies prioritizing actual over potential personhood, reinforce these delays but remain secondary to empirical ontogeny.[115] Critiques of delayed personhood emphasize the absence of empirical thresholds post-fertilization that fundamentally alter the zygote's organismal identity, as development proceeds via intrinsic genetic programming without discontinuous "emergence" of a new entity.[115] Viability, for instance, is critiqued as extrinsic and variable—advancing with neonatal technology from 28 weeks in the 1980s to under 24 weeks today—lacking causal grounding in the zygote's self-directed trajectory, which persists unchanged across stages.[116][109] Sentience-based claims similarly falter, as neural maturation builds on the pre-existing organism rather than conferring novel identity, with biological research affirming no post-zygotic event resets the human developmental continuum.[4][105] While philosophical personhood debates invoke values over strict biology, sources advancing delays often reflect institutional biases toward permissive frameworks, diverging from uncontroverted embryological data on fertilization as the origin of the organism.[111][109]References
- https://en.wiktionary.org/wiki/zygote