Recent from talks
Nothing was collected or created yet.
Transmembrane protein
View on Wikipedia
A transmembrane protein is a type of integral membrane protein that spans the entirety of the cell membrane. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane. They are usually highly hydrophobic and aggregate and precipitate in water. They require detergents or nonpolar solvents for extraction, although some of them (beta-barrels) can be also extracted using denaturing agents.
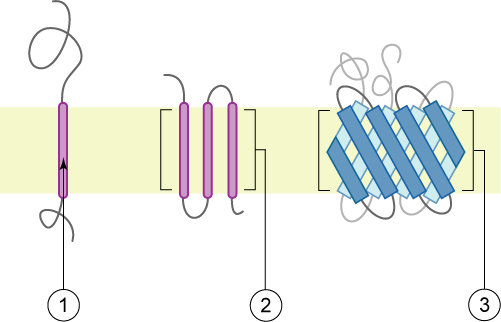
The peptide sequence that spans the membrane, or the transmembrane segment, is largely hydrophobic and can be visualized using the hydropathy plot.[1] Depending on the number of transmembrane segments, transmembrane proteins can be classified as single-pass membrane proteins, or as multipass membrane proteins.[2] Some other integral membrane proteins are called monotopic, meaning that they are also permanently attached to the membrane, but do not pass through it.[3]
Types
[edit]Classification by structure
[edit]There are two basic types of transmembrane proteins:[4] alpha-helical and beta barrels. Alpha-helical proteins are present in the inner membranes of bacterial cells or the plasma membrane of eukaryotic cells, and sometimes in the bacterial outer membrane.[5] This is the major category of transmembrane proteins. In humans, 27% of all proteins have been estimated to be alpha-helical membrane proteins.[6] Beta-barrel proteins are so far found only in outer membranes of gram-negative bacteria, cell walls of gram-positive bacteria, outer membranes of mitochondria and chloroplasts, or can be secreted as pore-forming toxins. All beta-barrel transmembrane proteins have simplest up-and-down topology, which may reflect their common evolutionary origin and similar folding mechanism.[7]
In addition to the protein domains, there are unusual transmembrane elements formed by peptides. A typical example is gramicidin A, a peptide that forms a dimeric transmembrane β-helix.[8] This peptide is secreted by gram-positive bacteria as an antibiotic. A transmembrane polyproline-II helix has not been reported in natural proteins. Nonetheless, this structure was experimentally observed in specifically designed artificial peptides.[9]
Classification by topology
[edit]This classification refers to the position of the protein N- and C-termini on the different sides of the lipid bilayer. Types I, II, III and IV are single-pass molecules. Type I transmembrane proteins are anchored to the lipid membrane with a stop-transfer anchor sequence and have their N-terminal domains targeted to the endoplasmic reticulum (ER) lumen during synthesis (and the extracellular space, if mature forms are located on cell membranes). Type II and III are anchored with a signal-anchor sequence, with type II being targeted to the ER lumen with its C-terminal domain, while type III have their N-terminal domains targeted to the ER lumen. Type IV is subdivided into IV-A, with their N-terminal domains targeted to the cytosol and IV-B, with an N-terminal domain targeted to the lumen.[10] The implications for the division in the four types are especially manifest at the time of translocation and ER-bound translation, when the protein has to be passed through the ER membrane in a direction dependent on the type.[citation needed]

3D structure
[edit]
Membrane protein structures can be determined by X-ray crystallography, electron microscopy or NMR spectroscopy.[12] The most common tertiary structures of these proteins are transmembrane helix bundle and beta barrel. The portion of the membrane proteins that are attached to the lipid bilayer (see annular lipid shell) consist mostly of hydrophobic amino acids.[13]
Membrane proteins which have hydrophobic surfaces, are relatively flexible and are expressed at relatively low levels. This creates difficulties in obtaining enough protein and then growing crystals. Hence, despite the significant functional importance of membrane proteins, determining atomic resolution structures for these proteins is more difficult than globular proteins.[14] As of January 2013 less than 0.1% of protein structures determined were membrane proteins despite being 20–30% of the total proteome.[15] Due to this difficulty and the importance of this class of proteins methods of protein structure prediction based on hydropathy plots, the positive inside rule and other methods have been developed.[16][17][18]
Thermodynamic stability and folding
[edit]Stability of alpha-helical transmembrane proteins
[edit]Transmembrane alpha-helical (α-helical) proteins are unusually stable judging from thermal denaturation studies, because they do not unfold completely within the membranes (the complete unfolding would require breaking down too many α-helical H-bonds in the nonpolar media). On the other hand, these proteins easily misfold, due to non-native aggregation in membranes, transition to the molten globule states, formation of non-native disulfide bonds, or unfolding of peripheral regions and nonregular loops that are locally less stable.[19]
It is also important to properly define the unfolded state. The unfolded state of membrane proteins in detergent micelles is different from that in the thermal denaturation experiments.[citation needed] This state represents a combination of folded hydrophobic α-helices and partially unfolded segments covered by the detergent. For example, the "unfolded" bacteriorhodopsin in SDS micelles has four transmembrane α-helices folded, while the rest of the protein is situated at the micelle-water interface and can adopt different types of non-native amphiphilic structures. Free energy differences between such detergent-denatured and native states are similar to stabilities of water-soluble proteins (< 10 kcal/mol).[citation needed]
Folding of α-helical transmembrane proteins
[edit]Refolding of α-helical transmembrane proteins in vitro is technically difficult. There are relatively few examples of the successful refolding experiments, as for bacteriorhodopsin. In vivo, all such proteins are normally folded co-translationally within the large transmembrane translocon. The translocon channel provides a highly heterogeneous environment for the nascent transmembrane α-helices. A relatively polar amphiphilic α-helix can adopt a transmembrane orientation in the translocon (although it would be at the membrane surface or unfolded in vitro), because its polar residues can face the central water-filled channel of the translocon. Such mechanism is necessary for incorporation of polar α-helices into structures of transmembrane proteins. The amphiphilic helices remain attached to the translocon until the protein is completely synthesized and folded. If the protein remains unfolded and attached to the translocon for too long, it is degraded by specific "quality control" cellular systems.[citation needed]
Stability and folding of beta-barrel transmembrane proteins
[edit]Stability of beta barrel (β-barrel) transmembrane proteins is similar to stability of water-soluble proteins, based on chemical denaturation studies. Some of them are very stable even in chaotropic agents and high temperature. Their folding in vivo is facilitated by water-soluble chaperones, such as protein Skp. It is thought that β-barrel membrane proteins come from one ancestor even having different number of sheets which could be added or doubled during evolution. Some studies show a huge sequence conservation among different organisms and also conserved amino acids which hold the structure and help with folding.[20]
3D structures
[edit]Light absorption-driven transporters
[edit]- Bacteriorhodopsin-like proteins including rhodopsin (see also opsin)
- Bacterial photosynthetic reaction centres and photosystems I and II
- Light-harvesting complexes from bacteria and chloroplasts
Oxidoreduction-driven transporters
[edit]- Transmembrane cytochrome b-like proteins: coenzyme Q - cytochrome c reductase (cytochrome bc1 ); cytochrome b6f complex; formate dehydrogenase, respiratory nitrate reductase; succinate - coenzyme Q reductase (fumarate reductase); and succinate dehydrogenase. See electron transport chain.
- Cytochrome c oxidases from bacteria and mitochondria
Electrochemical potential-driven transporters
[edit]- Proton or sodium translocating F-type and V-type ATPases
P-P-bond hydrolysis-driven transporters
[edit]- P-type calcium ATPase (five different conformations)
- Calcium ATPase regulators phospholamban and sarcolipin
- ABC transporters
- General secretory pathway (Sec) translocon (preprotein translocase SecY)
Porters (uniporters, symporters, antiporters)
[edit]- Mitochondrial carrier proteins
- Major Facilitator Superfamily (Glycerol-3-phosphate transporter, Lactose permease, and Multidrug transporter EmrD)
- Resistance-nodulation-cell division (multidrug efflux transporter AcrB, see multidrug resistance)
- Dicarboxylate/amino acid:cation symporter (proton glutamate symporter)
- Monovalent cation/proton antiporter (Sodium/proton antiporter 1 NhaA)
- Neurotransmitter sodium symporter
- Ammonia transporters
- Drug/Metabolite Transporter (small multidrug resistance transporter EmrE - the structures are retracted as erroneous)
Alpha-helical channels including ion channels
[edit]- Voltage-gated ion channel like, including potassium channels KcsA and KvAP, and inward-rectifier potassium ion channel Kirbac
- Large-conductance mechanosensitive channel, MscL
- Small-conductance mechanosensitive ion channel (MscS)
- CorA metal ion transporters
- Ligand-gated ion channel of neurotransmitter receptors (acetylcholine receptor)
- Aquaporins
- Chloride channels
- Outer membrane auxiliary proteins (polysaccharide transporter) - α-helical transmembrane proteins from the outer bacterial membrane
Enzymes
[edit]- Methane monooxygenase
- Rhomboid protease
- Disulfide bond formation protein (DsbA-DsbB complex)
Proteins with single transmembrane alpha-helices
[edit]- Subunits of T cell receptor complex
- Cytochrome c nitrite reductase complex
- Glycophorin A dimer
- Inovirus (filamentous phage) major coat protein
- Pilin
- Pulmonary surfactant-associated protein
- Monoamine oxidases A and B
- Fatty acid amide hydrolase[21]
- Cytochrome P450 oxidases
- Corticosteroid 11β-dehydrogenases .
- Signal Peptide Peptidase
Beta-barrels composed of a single polypeptide chain
[edit]- Beta barrels from eight beta-strands and with "shear number" of ten (n=8, S=10). They include:
- Autotransporter domain (n=12,S=14)
- FadL outer membrane protein transport family, including Fatty acid transporter FadL (n=14,S=14)
- General bacterial porin family, known as trimeric porins (n=16,S=20)
- Maltoporin, or sugar porins (n=18,S=22)
- Nucleoside-specific porin (n=12,S=16)
- Outer membrane phospholipase A1(n=12,S=16)
- TonB-dependent receptors and their plug domain. They are ligand-gated outer membrane channels (n=22,S=24), including cobalamin transporter BtuB, Fe(III)-pyochelin receptor FptA, receptor FepA, ferric hydroxamate uptake receptor FhuA, transporter FecA, and pyoverdine receptor FpvA
- Outer membrane protein OpcA family (n=10,S=12) that includes outer membrane protease OmpT and adhesin/invasin OpcA protein
- Outer membrane protein G porin family (n=14,S=16)
Note: n and S are, respectively, the number of beta-strands and the "shear number"[22] of the beta-barrel
Beta-barrels composed of several polypeptide chains
[edit]- Trimeric autotransporter (n=12,S=12)
- Outer membrane efflux proteins, also known as trimeric outer membrane factors (n=12,S=18) including TolC and multidrug resistance proteins
- MspA porin (octamer, n=S=16) and α-hemolysin (heptamer n=S=14) . These proteins are secreted.
See also
[edit]References
[edit]- ^ Manor, Joshua; Feldblum, Esther S.; Arkin, Isaiah T. (2012). "Environment Polarity in Proteins Mapped Noninvasively by FTIR Spectroscopy". The Journal of Physical Chemistry Letters. 3 (7): 939–944. doi:10.1021/jz300150v. PMC 3341589. PMID 22563521.
- ^ Alberts, Bruce; Johnson, Alexander; Lewis, Julian; Raff, Martin; Roberts, Keith; Walter, Peter (2002). "Membrane Proteins". Molecular Biology of the Cell. 4th edition. Garland Science. Retrieved 31 October 2023.
- ^ Steven R. Goodman (2008). Medical cell biology. Academic Press. pp. 37–. ISBN 978-0-12-370458-0. Retrieved 24 November 2010.
- ^ Jin Xiong (2006). Essential bioinformatics. Cambridge University Press. pp. 208–. ISBN 978-0-521-84098-9. Retrieved 13 November 2010.
- ^ alpha-helical proteins in outer membranes include Stannin and certain lipoproteins, and others
- ^ Almén MS, Nordström KJ, Fredriksson R, Schiöth HB (2009). "Mapping the human membrane proteome: a majority of the human membrane proteins can be classified according to function and evolutionary origin". BMC Biol. 7: 50. doi:10.1186/1741-7007-7-50. PMC 2739160. PMID 19678920.
- ^ "Types of Transmembrane Proteins". WikiDoc. Retrieved 30 January 2025.
- ^ Nicholson, L. K.; Cross, T. A. (1989). "Gramicidin cation channel: an experimental determination of the right-handed helix sense and verification of .beta.-type hydrogen bonding". Biochemistry. 28 (24): 9379–9385. doi:10.1021/bi00450a019. PMID 2482072.
- ^ Kubyshkin, Vladimir; Grage, Stephan L.; Ulrich, Anne S.; Budisa, Nediljko (2019). "Bilayer thickness determines the alignment of model polyproline helices in lipid membranes". Physical Chemistry Chemical Physics. 21 (40): 22396–22408. Bibcode:2019PCCP...2122396K. doi:10.1039/c9cp02996f. PMID 31577299.
- ^ Harvey Lodish etc.; Molecular Cell Biology, Sixth edition, p.546
- ^ Goder, Veit; Spiess, Martin (31 August 2001). "Topogenesis of membrane proteins: determinants and dynamics". FEBS Letters. 504 (3): 87–93. doi:10.1016/S0014-5793(01)02712-0. PMID 11532438.
- ^ Cross, Timothy A.; Sharma, Mukesh; Yi, Myunggi; Zhou, Huan-Xiang (2011). "Influence of Solubilizing Environments on Membrane Protein Structures". Trends in Biochemical Sciences. 36 (2): 117–125. doi:10.1016/j.tibs.2010.07.005. PMC 3161620. PMID 20724162.
- ^ White, Stephen. "General Principle of Membrane Protein Folding and Stability". Stephen White Laboratory Homepage. 10 Nov. 2009. web.[verification needed]
- ^ Carpenter, Elisabeth P; Beis, Konstantinos; Cameron, Alexander D; Iwata, So (October 2008). "Overcoming the challenges of membrane protein crystallography". Current Opinion in Structural Biology. 18 (5): 581–586. doi:10.1016/j.sbi.2008.07.001. PMC 2580798. PMID 18674618.
- ^ "Membrane Proteins of known 3D Structure". Archived from the original on 2013-12-25. Retrieved 2016-05-01.
- ^ Elofsson, Arne; Heijne, Gunnar von (7 June 2007). "Membrane Protein Structure: Prediction versus Reality". Annual Review of Biochemistry. 76 (1): 125–140. CiteSeerX 10.1.1.332.4023. doi:10.1146/annurev.biochem.76.052705.163539. PMID 17579561.
- ^ Chen, Chien Peter; Rost, Burkhard (2002). "State-of-the-art in membrane protein prediction". Applied Bioinformatics. 1 (1): 21–35. CiteSeerX 10.1.1.134.7424. PMID 15130854.
- ^ Hopf, Thomas A.; Colwell, Lucy J.; Sheridan, Robert; Rost, Burkhard; Sander, Chris; Marks, Debora S. (June 2012). "Three-Dimensional Structures of Membrane Proteins from Genomic Sequencing". Cell. 149 (7): 1607–1621. doi:10.1016/j.cell.2012.04.012. PMC 3641781. PMID 22579045.
- ^ "Stability of α-helical transmembrane proteins". WikiDoc. Retrieved 30 January 2025.
- ^ Michalik, Marcin; Orwick-Rydmark, Marcella; Habeck, Michael; Alva, Vikram; Arnold, Thomas; Linke, Dirk; Permyakov, Eugene A. (3 August 2017). "An evolutionarily conserved glycine-tyrosine motif forms a folding core in outer membrane proteins". PLOS ONE. 12 (8) e0182016. Bibcode:2017PLoSO..1282016M. doi:10.1371/journal.pone.0182016. PMC 5542473. PMID 28771529.
- ^ Bracey MH, Hanson MA, Masuda KR, Stevens RC, Cravatt BF (November 2002). "Structural adaptations in a membrane enzyme that terminates endocannabinoid signaling". Science. 298 (5599): 1793–6. Bibcode:2002Sci...298.1793B. doi:10.1126/science.1076535. PMID 12459591. S2CID 22656813.
- ^ Murzin AG, Lesk AM, Chothia C (March 1994). "Principles determining the structure of beta-sheet barrels in proteins. I. A theoretical analysis". J. Mol. Biol. 236 (5): 1369–81. doi:10.1016/0022-2836(94)90064-7. PMID 8126726.