Recent from talks
Nothing was collected or created yet.
Transcription factor II H
View on Wikipedia
| general transcription factor IIH, polypeptide 1, 62kDa | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Symbol | GTF2H1 | ||||||
| Alt. symbols | BTF2 | ||||||
| NCBI gene | 2965 | ||||||
| HGNC | 4655 | ||||||
| OMIM | 189972 | ||||||
| RefSeq | NM_005316 | ||||||
| UniProt | P32780 | ||||||
| Other data | |||||||
| Locus | Chr. 11 p15.1-p14 | ||||||
| |||||||
| general transcription factor IIH, polypeptide 2, 44kDa | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Symbol | GTF2H2 | ||||||
| Alt. symbols | BTF2, TFIIH, BTF2P44, T-BTF2P44 | ||||||
| NCBI gene | 2966 | ||||||
| HGNC | 4656 | ||||||
| OMIM | 601748 | ||||||
| RefSeq | NM_001515 | ||||||
| UniProt | Q13888 | ||||||
| Other data | |||||||
| Locus | Chr. 5 q12.2-13.3 | ||||||
| |||||||
| general transcription factor IIH, polypeptide 3, 34kDa | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Symbol | GTF2H3 | ||||||
| Alt. symbols | BTF2, TFIIH | ||||||
| NCBI gene | 2967 | ||||||
| HGNC | 4657 | ||||||
| OMIM | 601750 | ||||||
| RefSeq | NM_001516 | ||||||
| UniProt | Q13889 | ||||||
| Other data | |||||||
| Locus | Chr. 12 q24.31 | ||||||
| |||||||
Transcription factor II H (TFIIH) is a multi-subunit protein complex involved in both the transcription of protein-coding genes and the nucleotide excision repair (NER) pathway. TFIIH was first identified in 1989 as general transcription factor-δ or basic transcription factor 2, an essential factor for transcription in vitro. It was subsequently isolated from yeast and officially named TFIIH in 1992.[1][2]
TFIIH is composed of ten subunits. Seven of these—ERCC2/XPD, ERCC3/XPB, GTF2H1/p62, GTF2H4/p52, GTF2H2/p44, GTF2H3/p34, and GTF2H5/TTDA—constitute the core complex. The remaining three subunits—CDK7, MAT1, and cyclin H—form the cyclin-activating kinase (CAK) subcomplex, which is tethered to the core via the XPD protein.[3] Among the core subunits, ERCC2/XPD and ERCC3/XPB possess helicase and ATPase activities and are essential for unwinding DNA to form the transcription bubble. These activities are necessary during transcription in vitro only when the DNA template is not already denatured or is supercoiled.
The CAK subunits, CDK7 and cyclin H, are responsible for the phosphorylation of serine residues in the C-terminal domain of RNA polymerase II, as well as potentially other targets involved in the cell cycle. In addition to its essential role in transcription initiation, TFIIH also plays a critical part in nucleotide excision repair.
History
[edit]Before being designated as TFIIH, the complex was known by several names. It was first isolated in 1989 from rat liver and referred to as transcription factor δ. When identified in cancer cells, it was called basic transcription factor 2, and when isolated from yeast, it was known as transcription factor B. The complex was officially named TFIIH in 1992.[4]
Structure
[edit]TFIIH is a ten‐subunit complex; seven of these subunits comprise the "core" whereas three comprise the dissociable "CAK" (CDK-activating Kinase) module.[5] The core consists of subunits XPB, XPD, p62, p52, p44, p34 and p8 while CAK is composed of CDK7, cyclin H, and MAT1.[5]
Function
[edit]General functions of TFIIH include:
Gene transcription
[edit]TFIIH is a general transcription factor that helps recruit RNA polymerase II (Pol II) to gene promoters. It acts as a DNA translocase, sliding along the DNA while feeding it into the RNA polymerase II cleft, thereby generating torsional strain that facilitates local DNA unwinding.[7] TFIIH also plays a critical role in nucleotide excision repair (NER), where it unwinds DNA at sites of damage following lesion recognition by either the global genome repair (GGR) or transcription-coupled repair (TCR) pathway.[8][9]
DNA repair
[edit]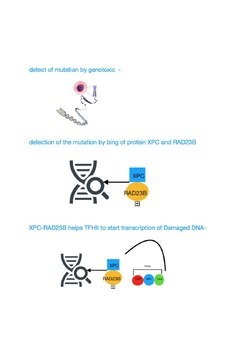
TFIIH participates in nucleotide excision repair (NER) by opening the DNA double helix after damage is initially recognized. NER is a multi-step pathway that removes a wide range of different types of damage that distort normal base pairing, including bulky chemical damage and UV-induced damage. Individuals with mutational defects in genes specifying protein components that catalyze the NER pathway, including the TFIIH components, often display features of premature aging.[10][11]
Clinical significance
[edit]Trichothiodystrophy
[edit]Mutation in genes ERCC3 (XPB), ERCC2 (XPD) or GTF2H5 (TTDA) cause trichothiodystrophy, a condition characterized by photosensitivity, ichthyosis, brittle hair and nails, intellectual impairment, decreased fertility and/or short stature.[10]
Cancer
[edit]Genetic polymorphisms of genes that encode subunits of TFIIH are known to be associated with increased cancer susceptibility in many tissues, e.g. skin tissue, breast tissue and lung tissue. Mutations in the subunits (such as XPD and XPB) can lead to a variety of diseases, including xeroderma pigmentosum (XP) or XP combined with Cockayne syndrome.[12]
Viral infection
[edit]Virus-encoded proteins target TFIIH.[13]
Inhibitors
[edit]Potent, bioactive natural products such as triptolide, which inhibit mammalian transcription by targeting the XPB subunit of the general transcription factor TFIIH, have recently been developed as glucose conjugates to selectively target hypoxic cancer cells with elevated glucose transporter expression.[14]
References
[edit]- ^ Flores O, Lu H, Reinberg D (February 1992). "Factors involved in specific transcription by mammalian RNA polymerase II. Identification and characterization of factor IIH". The Journal of Biological Chemistry. 267 (4): 2786–2793. doi:10.1016/S0021-9258(18)45947-9. PMID 1733973.
- ^ Kim TK, Ebright RH, Reinberg D (May 2000). "Mechanism of ATP-dependent promoter melting by transcription factor IIH". Science. 288 (5470). New York, N.Y.: 1418–1422. Bibcode:2000Sci...288.1418K. doi:10.1126/science.288.5470.1418. PMID 10827951.
- ^ Lee TI, Young RA (2000). "Transcription of eukaryotic protein-coding genes". Annual Review of Genetics. 34: 77–137. doi:10.1146/annurev.genet.34.1.77. PMID 11092823.
- ^ Rimel JK, Taatjes DJ (June 2018). "The essential and multifunctional TFIIH complex". Protein Science. 27 (6): 1018–1037. doi:10.1002/pro.3424. PMC 5980561. PMID 29664212.
- ^ a b Drapkin R, Reardon JT, Ansari A, Huang JC, Zawel L, Ahn K, et al. (April 1994). "Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II". Nature. 368 (6473): 769–772. Bibcode:1994Natur.368..769D. doi:10.1038/368769a0. PMID 8152490. S2CID 4363484.
- ^ a b Compe E, Egly JM (May 2012). "TFIIH: when transcription met DNA repair". Nature Reviews. Molecular Cell Biology. 13 (6): 343–354. doi:10.1038/nrm3350. PMID 22572993. S2CID 29077515.
- ^ Fishburn J, Tomko E, Galburt E, Hahn S (2015). "Double-stranded DNA translocase activity of transcription factor TFIIH and the mechanism of RNA polymerase II open complex formation". Proceedings of the National Academy of Sciences of the United States of America. 112 (13): 3961–3966. Bibcode:2015PNAS..112.3961F. doi:10.1073/pnas.1417709112. PMC 4386358. PMID 25775526.
- ^ Hoogstraten D, Nigg AL, Heath H, Mullenders LH, van Driel R, Hoeijmakers JH, et al. (November 2002). "Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo". Molecular Cell. 10 (5): 1163–1174. doi:10.1016/s1097-2765(02)00709-8. PMID 12453423.
- ^ Assfalg R, Lebedev A, Gonzalez OG, Schelling A, Koch S, Iben S (January 2012). "TFIIH is an elongation factor of RNA polymerase I". Nucleic Acids Research. 40 (2): 650–659. doi:10.1093/nar/gkr746. PMC 3258137. PMID 21965540.
- ^ a b Theil AF, Hoeijmakers JH, Vermeulen W (November 2014). "TTDA: big impact of a small protein". Experimental Cell Research. 329 (1): 61–68. doi:10.1016/j.yexcr.2014.07.008. PMID 25016283.
- ^ Edifizi D, Schumacher B (August 2015). "Genome Instability in Development and Aging: Insights from Nucleotide Excision Repair in Humans, Mice, and Worms". Biomolecules. 5 (3): 1855–1869. doi:10.3390/biom5031855. PMC 4598778. PMID 26287260.
- ^ Oh KS, Khan SG, Jaspers NG, Raams A, Ueda T, Lehmann A, et al. (November 2006). "Phenotypic heterogeneity in the XPB DNA helicase gene (ERCC3): xeroderma pigmentosum without and with Cockayne syndrome". Human Mutation. 27 (11): 1092–1103. doi:10.1002/humu.20392. PMID 16947863. S2CID 22852219.
- ^ Le May N, Dubaele S, Proietti De Santis L, Billecocq A, Bouloy M, Egly JM (February 2004). "TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus". Cell. 116 (4): 541–550. doi:10.1016/s0092-8674(04)00132-1. PMID 14980221. S2CID 14312462.
- ^ Datan E, Minn I, Peng X, He QL, Ahn H, Yu B, et al. (Sep 2020). "A Glucose-Triptolide Conjugate Selectively Targets Cancer Cells under Hypoxia". iScience. 23 (9) 101536. Bibcode:2020iSci...23j1536D. doi:10.1016/j.isci.2020.101536. PMC 7509213. PMID 33083765.
External links
[edit]- Transcription+Factor+TFIIH at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
Transcription factor II H
View on GrokipediaIntroduction
Overview
Transcription factor II H (TFIIH) is a heterodecameric protein complex that functions as a general transcription factor essential for the initiation of RNA polymerase II (Pol II)-dependent transcription in eukaryotes.[1] It plays critical roles not only in basal transcription but also in nucleotide excision repair (NER) of DNA damage and cell cycle regulation through its cyclin-dependent kinase (CDK)-activating kinase (CAK) module.[6] In transcription, TFIIH unwinds promoter DNA using its helicase subunits XPB and XPD, facilitating open complex formation and promoter clearance by Pol II.[7] The complex comprises 10 subunits organized into a core subcomplex of seven subunits (XPB, XPD, p62, p52, p44, p34, and p8) and a CAK subcomplex consisting of three subunits (CDK7, cyclin H, and MAT1), with the CAK loosely associated and sometimes dissociable.[8] This modular architecture enables TFIIH's multifunctional capabilities, allowing it to integrate signaling from transcription, repair, and cell cycle pathways.[1] Through its involvement in Pol II transcription, TFIIH is responsible for synthesizing all protein-coding messenger RNAs (mRNAs) and many noncoding RNAs in eukaryotic cells, accounting for the vast majority of gene expression.[9] TFIIH is evolutionarily conserved across eukaryotes, from yeast to humans, with its core subunits showing high sequence and functional similarity that underscores its fundamental role in cellular homeostasis.[10] In humans, mutations in TFIIH subunits are linked to genetic disorders such as xeroderma pigmentosum and Cockayne syndrome, highlighting its indispensability.[11] Studies on human TFIIH have revealed its dynamic conformational changes that coordinate these diverse activities.[2]Nomenclature
Transcription factor II H (TFIIH) is the established nomenclature for this eukaryotic multi-subunit protein complex essential for RNA polymerase II-mediated transcription initiation. The full designation reflects its role as a transcription factor specific to RNA polymerase II, with the "H" indicating its classification within the general transcription factor series (TFIIA through TFIIJ). The HUGO Gene Nomenclature Committee (HGNC) recognizes TFIIH as comprising subunits collectively termed general transcription factor IIH complex subunits, emphasizing their coordinated function in basal transcription machinery.[12] Synonyms for TFIIH include general transcription factor 2H, reflecting its basal role in promoter recognition and pre-initiation complex assembly. These alternative terms appear frequently in scientific literature to denote the complex's multifunctional nature, though TFIIH remains the primary identifier in modern nomenclature. Basal transcription factor is another occasional synonym, underscoring its necessity for RNA polymerase II-dependent gene expression across eukaryotes.[13][1] Subunit nomenclature follows HGNC-approved conventions, combining functional descriptors, molecular weights, and disease associations. For instance, the core helicase subunits are designated XPB (Xeroderma pigmentosum group B protein, encoded by ERCC3) and XPD (Xeroderma pigmentosum group D protein, encoded by ERCC2), reflecting their identification through complementation studies in DNA repair disorders. Structural subunits include p62 (encoded by GTF2H1, previously BTF2), p44 (GTF2H2), p34 (GTF2H3), p52 (GTF2H4), and p8 (GTF2H5, also known as TTDA for Trichothiodystrophy complementation group A). The cyclin-activating kinase (CAK) subcomplex subunits are named cyclin H (CCNH), cyclin-dependent kinase 7 (CDK7), and MNAT1 (MAT1). These names often derive from biochemical purification (e.g., "p" for apparent molecular weight in kilodaltons) or yeast orthologs (e.g., BTF2 from basic transcription factor 2).[12][13] Historically, TFIIH was first purified and named transcription factor δ (delta) in the late 1980s from rat liver extracts, where it was identified as a multifunctional initiation factor with ATPase and kinase activities required for RNA polymerase II promoter opening. This early designation shifted to the current TFIIH nomenclature in the early 1990s as its full composition and roles in both transcription and nucleotide excision repair were elucidated through genetic and biochemical studies in yeast and human systems. The transition aligned with standardized naming for general transcription factors, distinguishing TFIIH from other complexes like TFIIF or TFIIE.[14][15]History
Discovery
The discovery of Transcription factor II H (TFIIH) stemmed from pioneering biochemical studies on eukaryotic mRNA synthesis in the 1980s, particularly those conducted in Robert G. Roeder's laboratory at The Rockefeller University. In 1980, Roeder and colleagues developed an in vitro transcription system using soluble extracts from human HeLa cells and purified RNA polymerase II, demonstrating that multiple accessory protein factors were essential for accurate initiation at the promoter of the adenovirus major late gene. This seminal work revealed the complexity of the basal transcription apparatus and set the stage for systematic fractionation to identify individual components, shifting the field from crude extracts to defined systems.[16] Between 1983 and 1985, Roeder's group and others performed extensive chromatographic fractionation of HeLa and rat liver nuclear extracts, successfully resolving the general transcription factors required for RNA polymerase II-mediated basal transcription into distinct activities labeled TFIIA through TFIIF, with an additional heat-labile fraction initially termed factor G or δ. This fraction was indispensable for reconstituting accurate transcription but resisted further purification due to its multisubunit nature and instability. By 1989, Ron C. Conaway and Joan W. Conaway, working in Roeder's lab, achieved over 3,000-fold purification of this factor (named δ) from rat liver nuclear extracts, identifying it as a ~500 kDa complex essential for open complex formation and promoter opening in in vitro assays. Notably, the purified factor exhibited DNA-dependent ATPase activity strongly stimulated by TATA box-containing promoters, providing early evidence of its capacity for DNA unwinding during transcription initiation. Independently, Jean-Marc Egly's group at the Institut de Génétique et de Biologie Moléculaire purified a functionally equivalent multisubunit complex from HeLa cells, termed basic transcription factor 2 (BTF2), which complemented the rat δ activity and confirmed its universal role in human basal transcription. These parallel efforts established TFIIH (unifying the nomenclature for δ and BTF2) as a core general transcription factor.[17][17] Subsequent investigations in the early 1990s refined TFIIH's mechanistic contributions. In 1993, Hiroyuki Serizawa, along with the Conaways in Roeder's laboratory, demonstrated through kinetic analyses in reconstituted systems that TFIIH (factor δ) facilitates promoter clearance—the ATP-dependent transition from abortive initiation to processive elongation—via its ATPase/helicase activities, independent of its associated kinase function for RNA polymerase II C-terminal domain phosphorylation. This finding underscored TFIIH's active role beyond preinitiation complex assembly, linking its enzymatic properties directly to promoter escape and early elongation. These discoveries, grounded in rigorous biochemical fractionation and functional assays, positioned TFIIH as a pivotal multifunctional complex in RNA polymerase II transcription.Subunit identification
The identification of TFIIH subunits began in the early 1990s through genetic complementation studies in xeroderma pigmentosum (XP) cell lines, which revealed the core helicase subunits XPB (encoded by ERCC3) and XPD (encoded by ERCC2). The ERCC3 gene was cloned in 1990 by complementing the repair defect in an XP group B patient-derived cell line, identifying XPB as an ATP-dependent DNA helicase essential for nucleotide excision repair (NER). Subsequent work in 1993 demonstrated that XPB is a component of the basal transcription factor BTF2, later renamed TFIIH, establishing its dual role in transcription and repair. Similarly, the ERCC2 gene was cloned around the same period via complementation of XP group D cells, with XPD identified as the second helicase subunit of TFIIH in 1993, confirming its integration into the complex for both processes. Sequential cloning efforts followed, elucidating the remaining core subunits. The p62 subunit (encoded by GTF2H1) was cloned in 1992 as the 62-kDa component of BTF2/TFIIH through biochemical purification and cDNA isolation from HeLa cell extracts, revealing its role in stabilizing the complex.[18] The p44 subunit (GTF2H2) and p34 subunit (GTF2H3) were identified in 1994 via cDNA cloning and sequence analysis, with p44 showing homology to yeast Ssl1 and p34 exhibiting homology to a domain of Ssl1, confirming their presence in the TFIIH core through immunoprecipitation studies. The p52 subunit (GTF2H4) was cloned in 1997 as the fifth core subunit, with homology to yeast Tfb2, and its integration into TFIIH verified by co-immunoprecipitation with other subunits. The p8 subunit (also known as TTDA or GTF2H5) was discovered in 2004 through genetic analysis of trichothiodystrophy (TTD) patients, where mutations in this gene were linked to reduced TFIIH stability; early hints from TTD patient cell lines in the late 1990s suggested a small stabilizing factor, but formal identification came via complementation and mutation screening. This subunit acts as a repair-specific stabilizer, with patient-derived mutations causing TFIIH instability without affecting transcription as severely as repair. Parallel identification of the CAK subcomplex occurred in the mid-1990s. The kinase subunit CDK7 (previously MO15) was characterized in 1994 as part of TFIIH through co-purification with the core complex from HeLa cells, demonstrating its role in phosphorylating the RNA polymerase II C-terminal domain.[19] Cyclin H was cloned and associated with CDK7 in the same year, forming the core kinase module essential for TFIIH's transcriptional activity. MAT1 was identified in 1995 as the third CAK component, stabilizing the CDK7-Cyclin H assembly and bridging it to the TFIIH core via interactions with XPD.[20] A key milestone came in the mid-2000s with biochemical and genetic studies confirming TFIIH's heterodecameric composition, integrating the seven core subunits (XPB, XPD, p62, p44, p52, p34, p8) and the three CAK subunits (CDK7, Cyclin H, MAT1) into a stable 500-kDa assembly, resolving earlier uncertainties about subcomplex stoichiometry.[21]Composition
Core subunits
The core of Transcription factor II H (TFIIH) consists of seven stably associated subunits that form the minimal complex required for nucleotide excision repair (NER) and basal transcription activities, excluding the detachable CAK subcomplex. These subunits are XPB (encoded by ERCC3, 782 amino acids, ~89 kDa), an ATP-dependent DNA helicase with 3'→5' polarity that unwinds DNA to promote promoter opening during transcription initiation; XPD (ERCC2, 760 amino acids, ~87 kDa), a 5'→3' DNA helicase essential for damage verification in NER; p62 (GTF2H1, 548 amino acids, ~62 kDa), a scaffold protein that mediates interactions with TFIIE and stabilizes the core architecture; p52 (GTF2H4, 464 amino acids, ~52 kDa), which binds and stabilizes XPB; p44 (GTF2H2, 395 amino acids, ~44 kDa), containing an iron-sulfur cluster that regulates XPD helicase activity; p34 (GTF2H3, 294 amino acids, ~34 kDa), involved in DNA binding and core structural integrity; and p8 (GTF2H5, 71 amino acids, ~8 kDa), a stabilizer that bridges XPB and XPD interactions within the complex.[22]| Subunit | Gene | Size (aa, kDa) | Key Properties |
|---|---|---|---|
| XPB | ERCC3 | 782, ~89 | ATPase/helicase (3'→5'), promoter unwinding |
| XPD | ERCC2 | 760, ~87 | Helicase (5'→3'), damage sensing in NER |
| p62 | GTF2H1 | 548, ~62 | Scaffold, TFIIE interaction |
| p52 | GTF2H4 | 464, ~52 | XPB stabilizer and recruiter |
| p44 | GTF2H2 | 395, ~44 | Fe-S cluster, XPD regulator |
| p34 | GTF2H3 | 294, ~34 | DNA binding, structural support |
| p8 | GTF2H5 | 71, ~8 | Core stabilizer, XPB/XPD bridging |
