Recent from talks
Nothing was collected or created yet.
Biochemistry
View on Wikipedia| Part of a series on |
| Biochemistry |
|---|
 |
This article needs additional citations for verification. (August 2025) |
Biochemistry, or biological chemistry, is the study of chemical processes within and relating to living organisms.[1] A sub-discipline of both chemistry and biology, biochemistry may be divided into three fields: structural biology, enzymology, and metabolism. Over the last decades of the 20th century, biochemistry has become successful at explaining living processes through these three disciplines. Almost all areas of the life sciences are being uncovered and developed through biochemical methodology and research.[2] Biochemistry focuses on understanding the chemical basis that allows biological molecules to give rise to the processes that occur within living cells and between cells,[3] in turn relating greatly to the understanding of tissues and organs as well as organism structure and function.[4] Biochemistry is closely related to molecular biology, the study of the molecular mechanisms of biological phenomena.[5]
Much of biochemistry deals with the structures, functions, and interactions of biological macromolecules such as proteins, nucleic acids, carbohydrates, and lipids. They provide the structure of cells and perform many of the functions associated with life.[6] The chemistry of the cell also depends upon the reactions of small molecules and ions. These can be inorganic (for example, water and metal ions) or organic (for example, the amino acids, which are used to synthesize proteins).[7] The mechanisms used by cells to harness energy from their environment via chemical reactions are known as metabolism. The findings of biochemistry are applied primarily in medicine, nutrition, and agriculture. In medicine, biochemists investigate the causes and cures of diseases.[8] Nutrition studies how to maintain health and wellness and also the effects of nutritional deficiencies.[9] In agriculture, biochemists investigate soil and fertilizers with the goal of improving crop cultivation, crop storage, and pest control. In recent decades, biochemical principles and methods have been combined with problem-solving approaches from engineering to manipulate living systems in order to produce useful tools for research, industrial processes, and diagnosis and control of disease—the discipline of biotechnology.
History
[edit]
At its most comprehensive definition, biochemistry can be seen as a study of the components and composition of living things and how they come together to become life. In this sense, the history of biochemistry may therefore go back as far as the ancient Greeks.[10] However, biochemistry as a specific scientific discipline began sometime in the 19th century, or a little earlier, depending on which aspect of biochemistry is being focused on. Some argued that the beginning of biochemistry may have been the discovery of the first enzyme, diastase (now called amylase), in 1833 by Anselme Payen,[11] while others considered Eduard Buchner's first demonstration of a complex biochemical process alcoholic fermentation in cell-free extracts in 1897 to be the birth of biochemistry.[12][13] Some might also point as its beginning to the influential 1842 work by Justus von Liebig, Animal chemistry, or, Organic chemistry in its applications to physiology and pathology, which presented a chemical theory of metabolism,[10] or even earlier to the 18th century studies on fermentation and respiration by Antoine Lavoisier.[14][15] Many other pioneers in the field who helped to uncover the layers of complexity of biochemistry have been proclaimed founders of modern biochemistry. Emil Fischer, who studied the chemistry of proteins,[16] and F. Gowland Hopkins, who studied enzymes and the dynamic nature of biochemistry, represent two examples of early biochemists.[17]
The term "biochemistry" was first used when Vinzenz Kletzinsky (1826–1882) had his "Compendium der Biochemie" printed in Vienna in 1858; it derived from a combination of biology and chemistry. In 1877, Felix Hoppe-Seyler used the term (biochemie in German) as a synonym for physiological chemistry in the foreword to the first issue of Zeitschrift für Physiologische Chemie (Journal of Physiological Chemistry) where he argued for the setting up of institutes dedicated to this field of study.[18][19] The German chemist Carl Neuberg however is often cited to have coined the word in 1903,[20][21][22] while some credited it to Franz Hofmeister.[23]

It was once generally believed that life and its materials had some essential property or substance (often referred to as the "vital principle") distinct from any found in non-living matter, and it was thought that only living beings could produce the molecules of life.[25] In 1828, Friedrich Wöhler published a paper on his serendipitous urea synthesis from potassium cyanate and ammonium sulfate; some regarded that as a direct overthrow of vitalism and the establishment of organic chemistry.[26][27] However, the Wöhler synthesis has sparked controversy as some reject the death of vitalism at his hands.[28] Since then, biochemistry has advanced, especially since the mid-20th century, with the development of new techniques such as chromatography, X-ray diffraction, dual polarisation interferometry, NMR spectroscopy, radioisotopic labeling, electron microscopy and molecular dynamics simulations. These techniques allowed for the discovery and detailed analysis of many molecules and metabolic pathways of the cell, such as glycolysis and the Krebs cycle (citric acid cycle), and led to an understanding of biochemistry on a molecular level.[citation needed]
Another significant historic event in biochemistry is the discovery of the gene, and its role in the transfer of information in the cell. In the 1950s, James D. Watson, Francis Crick, Rosalind Franklin and Maurice Wilkins were instrumental in solving DNA structure and suggesting its relationship with the genetic transfer of information.[29] In 1958, George Beadle and Edward Tatum received the Nobel Prize for work in fungi showing that one gene produces one enzyme.[30] In 1988, Colin Pitchfork was the first person convicted of murder with DNA evidence, which led to the growth of forensic science.[31] More recently, Andrew Z. Fire and Craig C. Mello received the 2006 Nobel Prize for discovering the role of RNA interference (RNAi) in the silencing of gene expression.[32]
Starting materials: the chemical elements of life
[edit]
Around two dozen chemical elements are essential to various kinds of biological life. Most rare elements on Earth are not needed by life (exceptions being selenium and iodine),[33] while a few common ones (aluminium and titanium) are not used. Most organisms share element needs, but there are a few differences between plants and animals. For example, ocean algae use bromine, but land plants and animals do not seem to need any. All animals require sodium, but is not an essential element for plants. Plants need boron and silicon, but animals may not (or may need ultra-small amounts).[34]
Just six elements—carbon, hydrogen, nitrogen, oxygen, calcium and phosphorus—make up almost 99% of the mass of living cells, including those in the human body (see composition of the human body for a complete list). In addition to the six major elements that compose most of the human body, humans require smaller amounts of possibly 18 more.[35]
Biomolecules
[edit]The 4 main classes of molecules in biochemistry (often called biomolecules) are carbohydrates, lipids, proteins, and nucleic acids.[36] Many biological molecules are polymers: in this terminology, monomers are relatively small macromolecules that are linked together to create large macromolecules known as polymers. When monomers are linked together to synthesize a biological polymer, they undergo a process called dehydration synthesis. Different macromolecules can assemble in larger complexes, often needed for biological activity.
Carbohydrates
[edit]Two of the main functions of carbohydrates are energy storage and providing structure. One of the common sugars known as glucose is a carbohydrate, but not all carbohydrates are sugars. There are more carbohydrates on Earth than any other known type of biomolecule; they are used to store energy and genetic information, as well as play important roles in cell to cell interactions and communications.[citation needed]
The simplest type of carbohydrate is a monosaccharide, which among other properties contains carbon, hydrogen, and oxygen, mostly in a ratio of 1:2:1 (generalized formula CnH2nOn, where n is at least 3). Glucose (C6H12O6) is one of the most important carbohydrates; others include fructose (C6H12O6), the sugar commonly associated with the sweet taste of fruits,[37][a] and deoxyribose (C5H10O4), a component of DNA. A monosaccharide can switch between acyclic (open-chain) form and a cyclic form. The open-chain form can be turned into a ring of carbon atoms bridged by an oxygen atom created from the carbonyl group of one end and the hydroxyl group of another. The cyclic molecule has a hemiacetal or hemiketal group, depending on whether the linear form was an aldose or a ketose.[39]
In these cyclic forms, the ring usually has 5 or 6 atoms. These forms are called furanoses and pyranoses, respectively—by analogy with furan and pyran, the simplest compounds with the same carbon-oxygen ring (although they lack the carbon-carbon double bonds of these two molecules). For example, the aldohexose glucose may form a hemiacetal linkage between the hydroxyl on carbon 1 and the oxygen on carbon 4, yielding a molecule with a 5-membered ring, called glucofuranose. The same reaction can take place between carbons 1 and 5 to form a molecule with a 6-membered ring, called glucopyranose. Cyclic forms with a 7-atom ring called heptoses are rare.[citation needed]
Two monosaccharides can be joined by a glycosidic or ester bond into a disaccharide through a dehydration reaction during which a molecule of water is released. The reverse reaction in which the glycosidic bond of a disaccharide is broken into two monosaccharides is termed hydrolysis. The best-known disaccharide is sucrose or ordinary sugar, which consists of a glucose molecule and a fructose molecule joined. Another important disaccharide is lactose found in milk, consisting of a glucose molecule and a galactose molecule. Lactose may be hydrolysed by lactase, and deficiency in this enzyme results in lactose intolerance.
When a few (around three to six) monosaccharides are joined, it is called an oligosaccharide (oligo- meaning "few"). These molecules tend to be used as markers and signals, as well as having some other uses.[40] Many monosaccharides joined form a polysaccharide. They can be joined in one long linear chain, or they may be branched. Two of the most common polysaccharides are cellulose and glycogen, both consisting of repeating glucose monomers. Cellulose is an important structural component of plant's cell walls and glycogen is used as a form of energy storage in animals.
Sugar can be characterized by having reducing or non-reducing ends. A reducing end of a carbohydrate is a carbon atom that can be in equilibrium with the open-chain aldehyde (aldose) or keto form (ketose). If the joining of monomers takes place at such a carbon atom, the free hydroxy group of the pyranose or furanose form is exchanged with an OH-side-chain of another sugar, yielding a full acetal. This prevents opening of the chain to the aldehyde or keto form and renders the modified residue non-reducing. Lactose contains a reducing end at its glucose moiety, whereas the galactose moiety forms a full acetal with the C4-OH group of glucose. Saccharose does not have a reducing end because of full acetal formation between the aldehyde carbon of glucose (C1) and the keto carbon of fructose (C2).
Lipids
[edit]
Lipids comprise a diverse range of molecules and to some extent is a catchall for relatively water-insoluble or nonpolar compounds of biological origin, including waxes, fatty acids, fatty-acid derived phospholipids, sphingolipids, glycolipids, and terpenoids (e.g., retinoids and steroids). Some lipids are linear, open-chain aliphatic molecules, while others have ring structures. Some are aromatic (with a cyclic [ring] and planar [flat] structure) while others are not. Some are flexible, while others are rigid.[43]
Lipids are usually made from one molecule of glycerol combined with other molecules. In triglycerides, the main group of bulk lipids, there is one molecule of glycerol and three fatty acids. Fatty acids are considered the monomer in that case, and may be saturated (no double bonds in the carbon chain) or unsaturated (one or more double bonds in the carbon chain).[citation needed]
Most lipids have some polar character and are largely nonpolar. In general, the bulk of their structure is nonpolar or hydrophobic ("water-fearing"), meaning that it does not interact well with polar solvents like water. Another part of their structure is polar or hydrophilic ("water-loving") and will tend to associate with polar solvents like water. This makes them amphiphilic molecules (having both hydrophobic and hydrophilic portions). In the case of cholesterol, the polar group is a mere –OH (hydroxyl or alcohol).[44]
In the case of phospholipids, the polar groups are considerably larger and more polar, as described below.
Lipids are an integral part of our daily diet. Most oils and milk products that we use for cooking and eating like butter, cheese, ghee etc. are composed of fats. Vegetable oils are rich in various polyunsaturated fatty acids (PUFA). Lipid-containing foods undergo digestion within the body and are broken into fatty acids and glycerol, the final degradation products of fats and lipids. Lipids, especially phospholipids, are also used in various pharmaceutical products, either as co-solubilizers (e.g. in parenteral infusions) or else as drug carrier components (e.g. in a liposome or transfersome).
Proteins
[edit]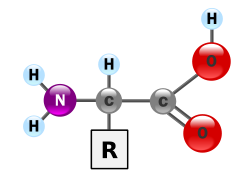
Proteins are very large molecules—macro-biopolymers—made from monomers called amino acids. An amino acid consists of an alpha carbon atom attached to an amino group, –NH2, a carboxylic acid group, –COOH (although these exist as –NH3+ and –COO− under physiologic conditions), a simple hydrogen atom, and a side chain commonly denoted as "–R". The side chain "R" is different for each amino acid of which there are 20 standard ones. It is this "R" group that makes each amino acid different, and the properties of the side chains greatly influence the overall three-dimensional conformation of a protein. Some amino acids have functions by themselves or in a modified form; for instance, glutamate functions as an important neurotransmitter. Amino acids can be joined via a peptide bond. In this dehydration synthesis, a water molecule is removed and the peptide bond connects the nitrogen of one amino acid's amino group to the carbon of the other's carboxylic acid group. The resulting molecule is called a dipeptide, and short stretches of amino acids (usually, fewer than thirty) are called peptides or polypeptides. Longer stretches merit the title proteins. As an example, the important blood serum protein albumin contains 585 amino acid residues.[45]

Proteins can have structural and/or functional roles. For instance, movements of the proteins actin and myosin ultimately are responsible for the contraction of skeletal muscle. One property many proteins have is that they specifically bind to a certain molecule or class of molecules—they may be extremely selective in what they bind. Antibodies are an example of proteins that attach to one specific type of molecule. Antibodies are composed of heavy and light chains. Two heavy chains would be linked to two light chains through disulfide linkages between their amino acids. Antibodies are specific through variation based on differences in the N-terminal domain.[46]
The enzyme-linked immunosorbent assay (ELISA), which uses antibodies, is one of the most sensitive tests modern medicine uses to detect various biomolecules. Probably the most important proteins, however, are the enzymes. Virtually every reaction in a living cell requires an enzyme to lower the activation energy of the reaction. These molecules recognize specific reactant molecules called substrates; they then catalyze the reaction between them. By lowering the activation energy, the enzyme speeds up that reaction by a rate of 1011 or more; a reaction that would normally take over 3,000 years to complete spontaneously might take less than a second with an enzyme. The enzyme itself is not used up in the process and is free to catalyze the same reaction with a new set of substrates. Using various modifiers, the activity of the enzyme can be regulated, enabling control of the biochemistry of the cell as a whole.
The structure of proteins is traditionally described in a hierarchy of four levels. The primary structure of a protein consists of its linear sequence of amino acids; for instance, "alanine-glycine-tryptophan-serine-glutamate-asparagine-glycine-lysine-...". Secondary structure is concerned with local morphology (morphology being the study of structure). Some combinations of amino acids will tend to curl up in a coil called an α-helix or into a sheet called a β-sheet; some α-helixes can be seen in the hemoglobin schematic above. Tertiary structure is the entire three-dimensional shape of the protein. This shape is determined by the sequence of amino acids. In fact, a single change can change the entire structure. The alpha chain of hemoglobin contains 146 amino acid residues; substitution of the glutamate residue at position 6 with a valine residue changes the behavior of hemoglobin so much that it results in sickle-cell disease. Finally, quaternary structure is concerned with the structure of a protein with multiple peptide subunits, like hemoglobin with its four subunits. Not all proteins have more than one subunit.[47]


Ingested proteins are usually broken up into single amino acids or dipeptides in the small intestine and then absorbed. They can then be joined to form new proteins. Intermediate products of glycolysis, the citric acid cycle, and the pentose phosphate pathway can be used to form all twenty amino acids, and most bacteria and plants possess all the necessary enzymes to synthesize them. Humans and other mammals, however, can synthesize only half of them. They cannot synthesize isoleucine, leucine, lysine, methionine, phenylalanine, threonine, tryptophan, and valine. Because they must be ingested, these are the essential amino acids. Mammals do possess the enzymes to synthesize alanine, asparagine, aspartate, cysteine, glutamate, glutamine, glycine, proline, serine, and tyrosine, the nonessential amino acids. While they can synthesize arginine and histidine, they cannot produce it in sufficient amounts for young, growing animals, and so these are often considered essential amino acids.
If the amino group is removed from an amino acid, it leaves behind a carbon skeleton called an α-keto acid. Enzymes called transaminases can easily transfer the amino group from one amino acid (making it an α-keto acid) to another α-keto acid (making it an amino acid). This is important in the biosynthesis of amino acids, as for many of the pathways, intermediates from other biochemical pathways are converted to the α-keto acid skeleton, and then an amino group is added, often via transamination. The amino acids may then be linked together to form a protein.
A similar process is used to break down proteins. It is first hydrolyzed into its component amino acids. Free ammonia (NH3), existing as the ammonium ion (NH4+) in blood, is toxic to life forms. A suitable method for excreting it must therefore exist. Different tactics have evolved in different animals, depending on the animals' needs. Unicellular organisms release the ammonia into the environment. Likewise, bony fish can release ammonia into the water where it is quickly diluted. In general, mammals convert ammonia into urea, via the urea cycle.
In order to determine whether two proteins are related, or in other words to decide whether they are homologous or not, scientists use sequence-comparison methods. Methods like sequence alignments and structural alignments are powerful tools that help scientists identify homologies between related molecules. The relevance of finding homologies among proteins goes beyond forming an evolutionary pattern of protein families. By finding how similar two protein sequences are, we acquire knowledge about their structure and therefore their function.
Nucleic acids
[edit]
Nucleic acids, so-called because of their prevalence in cellular nuclei, is the generic name of the family of biopolymers. They are complex, high-molecular-weight biochemical macromolecules that can convey genetic information in all living cells and viruses.[2] The monomers are called nucleotides, and each consists of three components: a nitrogenous heterocyclic base (either a purine or a pyrimidine), a pentose sugar, and a phosphate group.[48]

The most common nucleic acids are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). The phosphate group and the sugar of each nucleotide bond with each other to form the backbone of the nucleic acid, while the sequence of nitrogenous bases stores the information. The most common nitrogenous bases are adenine, cytosine, guanine, thymine, and uracil. The nitrogenous bases of each strand of a nucleic acid will form hydrogen bonds with certain other nitrogenous bases in a complementary strand of nucleic acid. Adenine binds with thymine and uracil, thymine binds only with adenine, and cytosine and guanine can bind only with one another. Adenine, thymine, and uracil contain two hydrogen bonds, while hydrogen bonds formed between cytosine and guanine are three.
Aside from the genetic material of the cell, nucleic acids often play a role as second messengers, as well as forming the base molecule for adenosine triphosphate (ATP), the primary energy-carrier molecule found in all living organisms. Also, the nitrogenous bases possible in the two nucleic acids are different: adenine, cytosine, and guanine occur in both RNA and DNA, while thymine occurs only in DNA and uracil occurs in RNA.
Metabolism
[edit]Carbohydrates as energy source
[edit]Glucose is an energy source in most life forms. For instance, polysaccharides are broken down into their monomers by enzymes (glycogen phosphorylase removes glucose residues from glycogen, a polysaccharide). Disaccharides like lactose or sucrose are cleaved into their two component monosaccharides.[49]
Glycolysis (anaerobic)
[edit]Glucose is mainly metabolized by a very important ten-step pathway called glycolysis, the net result of which is to break down one molecule of glucose into two molecules of pyruvate. This also produces a net two molecules of ATP, the energy currency of cells, along with two reducing equivalents of converting NAD+ (nicotinamide adenine dinucleotide: oxidized form) to NADH (nicotinamide adenine dinucleotide: reduced form). This does not require oxygen; if no oxygen is available (or the cell cannot use oxygen), the NAD is restored by converting the pyruvate to lactate (lactic acid) (e.g. in humans) or to ethanol plus carbon dioxide (e.g. in yeast). Other monosaccharides like galactose and fructose can be converted into intermediates of the glycolytic pathway.[50]
Aerobic
[edit]In aerobic cells with sufficient oxygen, as in most human cells, the pyruvate is further metabolized. It is irreversibly converted to acetyl-CoA, giving off one carbon atom as the waste product carbon dioxide, generating another reducing equivalent as NADH. The two molecules acetyl-CoA (from one molecule of glucose) then enter the citric acid cycle, producing two molecules of ATP, six more NADH molecules and two reduced (ubi)quinones (via FADH2 as enzyme-bound cofactor), and releasing the remaining carbon atoms as carbon dioxide. The produced NADH and quinol molecules then feed into the enzyme complexes of the respiratory chain, an electron transport system transferring the electrons ultimately to oxygen and conserving the released energy in the form of a proton gradient over a membrane (inner mitochondrial membrane in eukaryotes). Thus, oxygen is reduced to water and the original electron acceptors NAD+ and quinone are regenerated. This is why humans breathe in oxygen and breathe out carbon dioxide. The energy released from transferring the electrons from high-energy states in NADH and quinol is conserved first as proton gradient and converted to ATP via ATP synthase. This generates an additional 28 molecules of ATP (24 from the 8 NADH + 4 from the 2 quinols), totaling to 32 molecules of ATP conserved per degraded glucose (two from glycolysis + two from the citrate cycle).[51] It is clear that using oxygen to completely oxidize glucose provides an organism with far more energy than any oxygen-independent metabolic feature, and this is thought to be the reason why complex life appeared only after Earth's atmosphere accumulated large amounts of oxygen.
Gluconeogenesis
[edit]In vertebrates, vigorously contracting skeletal muscles (during weightlifting or sprinting, for example) do not receive enough oxygen to meet the energy demand, and so they shift to anaerobic metabolism, converting glucose to lactate. The combination of glucose from noncarbohydrates origin, such as fat and proteins. This only happens when glycogen supplies in the liver are worn out. The pathway is a crucial reversal of glycolysis from pyruvate to glucose and can use many sources like amino acids, glycerol and Krebs Cycle. Large scale protein and fat catabolism usually occur when those suffer from starvation or certain endocrine disorders.[52] The liver regenerates the glucose, using a process called gluconeogenesis. This process is not quite the opposite of glycolysis, and actually requires three times the amount of energy gained from glycolysis (six molecules of ATP are used, compared to the two gained in glycolysis). Analogous to the above reactions, the glucose produced can then undergo glycolysis in tissues that need energy, be stored as glycogen (or starch in plants), or be converted to other monosaccharides or joined into di- or oligosaccharides. The combined pathways of glycolysis during exercise, lactate's crossing via the bloodstream to the liver, subsequent gluconeogenesis and release of glucose into the bloodstream is called the Cori cycle.[53]
Relationship to other "molecular-scale" biological sciences
[edit]This section has multiple issues. Please help improve it or discuss these issues on the talk page. (Learn how and when to remove these messages)
|

Researchers in biochemistry use specific techniques native to biochemistry, but increasingly combine these with techniques and ideas developed in the fields of genetics, molecular biology, and biophysics. There is not a defined line between these disciplines. Biochemistry studies the chemistry required for biological activity of molecules, molecular biology studies their biological activity, genetics studies their heredity, which happens to be carried by their genome. This is shown in the following schematic that depicts one possible view of the relationships between the fields:
- Biochemistry is the study of the chemical substances and vital processes occurring in live organisms. Biochemists focus heavily on the role, function, and structure of biomolecules. The study of the chemistry behind biological processes and the synthesis of biologically active molecules are applications of biochemistry. Biochemistry studies life at the atomic and molecular level.
- Genetics is the study of the effect of genetic differences in organisms. This can often be inferred by the absence of a normal component (e.g. one gene). The study of "mutants" – organisms that lack one or more functional components with respect to the so-called "wild type" or normal phenotype. Genetic interactions (epistasis) can often confound simple interpretations of such "knockout" studies.
- Molecular biology is the study of molecular underpinnings of the biological phenomena, focusing on molecular synthesis, modification, mechanisms and interactions. The central dogma of molecular biology, where genetic material is transcribed into RNA and then translated into protein, despite being oversimplified, still provides a good starting point for understanding the field. This concept has been revised in light of emerging novel roles for RNA.
- Chemical biology seeks to develop new tools based on small molecules that allow minimal perturbation of biological systems while providing detailed information about their function. Further, chemical biology employs biological systems to create non-natural hybrids between biomolecules and synthetic devices (for example emptied viral capsids that can deliver gene therapy or drug molecules).
See also
[edit]Lists
[edit]See also
[edit]- Astrobiology
- Biochemistry (journal)
- Biological Chemistry (journal)
- Biophysics
- Chemical ecology
- Computational biomodeling
- Dedicated bio-based chemical
- EC number
- Hypothetical types of biochemistry
- International Union of Biochemistry and Molecular Biology
- Metabolome
- Metabolomics
- Molecular biology
- Molecular medicine
- Plant biochemistry
- Proteolysis
- Small molecule
- Structural biology
- TCA cycle
Notes
[edit]- ^ Fructose is not the only sugar found in fruits. Glucose and sucrose are also found in varying quantities in various fruits, and sometimes exceed the fructose present. For example, 32% of the edible portion of a date is glucose, compared with 24% fructose and 8% sucrose. However, peaches contain more sucrose (6.66%) than they do fructose (0.93%) or glucose (1.47%).[38]
References
[edit]- ^ "Biological/Biochemistry". acs.org. Archived from the original on 2019-08-21. Retrieved 2016-01-04.
- ^ a b Voet (2005), p. 3.
- ^ Karp (2009), p. 2.
- ^ Miller (2012). p. 62.
- ^ Astbury (1961), p. 1124.
- ^ Eldra (2007), p. 45.
- ^ Marks (2012), Chapter 14.
- ^ Finkel (2009), pp. 1–4.
- ^ UNICEF (2010), pp. 61, 75.
- ^ a b Helvoort (2000), p. 81.
- ^ Hunter (2000), p. 75.
- ^ Hamblin (2005), p. 26.
- ^ Hunter (2000), pp. 96–98.
- ^ Berg (1980), pp. 1–2.
- ^ Holmes (1987), p. xv.
- ^ Feldman (2001), p. 206.
- ^ Rayner-Canham (2005), p. 136.
- ^ Ziesak (1999), p. 169.
- ^ Kleinkauf (1988), p. 116.
- ^ Ben-Menahem (2009), p. 2982.
- ^ Amsler (1986), p. 55.
- ^ Horton (2013), p. 36.
- ^ Kleinkauf (1988), p. 43.
- ^ Edwards (1992), pp. 1161–1173.
- ^ Fiske (1890), pp. 419–20.
- ^ Wöhler, F. (1828). "Ueber künstliche Bildung des Harnstoffs". Annalen der Physik und Chemie. 88 (2): 253–256. Bibcode:1828AnP....88..253W. doi:10.1002/andp.18280880206. ISSN 0003-3804.
- ^ Kauffman (2001), pp. 121–133.
- ^ Lipman, Timothy O. (August 1964). "Wohler's preparation of urea and the fate of vitalism". Journal of Chemical Education. 41 (8): 452. Bibcode:1964JChEd..41..452L. doi:10.1021/ed041p452. ISSN 0021-9584.
- ^ Tropp (2012), pp. 19–20.
- ^ Krebs (2012), p. 32.
- ^ Butler (2009), p. 5.
- ^ Chandan (2007), pp. 193–194.
- ^ Cox, Nelson, Lehninger (2008). Lehninger Principles of Biochemistry. Macmillan.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ Sheng, Huachun; Lei, Yuyan; Wei, Jing; Yang, Zhengming; Peng, Lianxin; Li, Wenbing; Liu, Yuan (2024). "Analogy of silicon and boron in plant nutrition". Frontiers in Plant Science. 15 1353706. Bibcode:2024FrPS...1553706S. doi:10.3389/fpls.2024.1353706. ISSN 1664-462X. PMC 10877001. PMID 38379945.
- ^ Nielsen (1999), pp. 283–303.
- ^ Slabaugh (2007), pp. 3–6.
- ^ Whiting (1970), pp. 1–31.
- ^ Whiting, G.C. (1970), p. 5.
- ^ Voet (2005), pp. 358–359.
- ^ Varki (1999), p. 17.
- ^ Stryer (2007), p. 328.
- ^ Voet (2005), Ch. 12 Lipids and Membranes.
- ^ Ahmed, Saba; Shah, Parini; Ahmed, Owais (2023), "Biochemistry, Lipids", StatPearls, Treasure Island (FL): StatPearls Publishing, PMID 30247827, retrieved 2023-11-30
- ^ PhD, Chris Masterjohn (2008-11-09). "Cholesterol's Hydroxyl Group". Harnessing the Power of Nutrients. Retrieved 2025-08-29.
- ^ Metzler (2001), p. 58.
- ^ Feige, Matthias J.; Hendershot, Linda M.; Buchner, Johannes (2010). "How antibodies fold". Trends in Biochemical Sciences. 35 (4): 189–198. doi:10.1016/j.tibs.2009.11.005. PMC 4716677. PMID 20022755.
- ^ Fromm and Hargrove (2012), pp. 35–51.
- ^ Saenger (1984), p. 84.
- ^ "Disaccharide". Encyclopedia Britannica. Archived from the original on 19 October 2023. Retrieved 14 October 2023.
- ^ Fromm and Hargrove (2012), pp. 163–180.
- ^ Voet (2005), Ch. 17 Glycolysis.
- ^ A Dictionary of Biology. Oxford University Press. 17 September 2015. ISBN 978-0-19-871437-8. Archived from the original on 10 July 2020. Retrieved 29 April 2020.
- ^ Fromm and Hargrove (2012), pp. 183–194.
Cited literature
[edit]- Amsler, Mark (1986). The Languages of Creativity: Models, Problem-solving, Discourse. University of Delaware Press. ISBN 978-0-87413-280-9. Archived from the original on 2023-10-28. Retrieved 2015-07-27.
- Astbury, W.T. (1961). "Molecular Biology or Ultrastructural Biology ?". Nature. 190 (4781): 1124. Bibcode:1961Natur.190.1124A. doi:10.1038/1901124a0. PMID 13684868. S2CID 4172248.
- Ben-Menahem, Ari (2009). Historical Encyclopedia of Natural and Mathematical Sciences. Springer. p. 2982. Bibcode:2009henm.book.....B. ISBN 978-3-540-68831-0.
- Burton, Feldman (2001). The Nobel Prize: A History of Genius, Controversy, and Prestige. Arcade Publishing. ISBN 978-1-55970-592-9.
- Butler, John M. (2009). Fundamentals of Forensic DNA Typing. Academic Press. ISBN 978-0-08-096176-7.
- Sen, Chandan K.; Roy, Sashwati (2007). "MiRNA: Licensed to Kill the Messenger". DNA and Cell Biology. 26 (4): 193–194. doi:10.1089/dna.2006.0567. PMID 17465885. S2CID 10665411.
- Clarence, Peter Berg (1980). The University of Iowa and Biochemistry from Their Beginnings. University of Iowa. ISBN 978-0-87414-014-9. Archived from the original on 2023-10-28. Retrieved 2015-07-27.
- Edwards, Karen J.; Brown, David G.; Spink, Neil; Skelly, Jane V.; Neidle, Stephen (1992). "Molecular structure of the B-DNA dodecamer d(CGCAAATTTGCG)2 an examination of propeller twist and minor-groove water structure at 2·2Åresolution". Journal of Molecular Biology. 226 (4): 1161–1173. doi:10.1016/0022-2836(92)91059-x. PMID 1518049.
- Eldra P. Solomon; Linda R. Berg; Diana W. Martin (2007). Biology, 8th Edition, International Student Edition. Thomson Brooks/Cole. ISBN 978-0-495-31714-2. Archived from the original on 2016-03-04.
- Fariselli, P.; Rossi, I.; Capriotti, E.; Casadio, R. (2006). "The WWWH of remote homolog detection: The state of the art". Briefings in Bioinformatics. 8 (2): 78–87. doi:10.1093/bib/bbl032. PMID 17003074.
- Fiske, John (1890). Outlines of Cosmic Philosophy Based on the Doctrines of Evolution, with Criticisms on the Positive Philosophy, Volume 1. Boston and New York: Houghton, Mifflin. Archived from the original on 28 October 2023. Retrieved 16 February 2015.
- Finkel, Richard; Cubeddu, Luigi; Clark, Michelle (2009). Lippincott's Illustrated Reviews: Pharmacology (4th ed.). Lippincott Williams & Wilkins. ISBN 978-0-7817-7155-9. Archived from the original on 2023-10-28. Retrieved 2020-06-05.
- Krebs, Jocelyn E.; Goldstein, Elliott S.; Lewin, Benjamin; Kilpatrick, Stephen T. (2012). Essential Genes. Jones & Bartlett Publishers. ISBN 978-1-4496-1265-8.
- Fromm, Herbert J.; Hargrove, Mark (2012). Essentials of Biochemistry. Springer. ISBN 978-3-642-19623-2. Archived from the original on 2023-10-28. Retrieved 2020-06-05.
- Hamblin, Jacob Darwin (2005). Science in the Early Twentieth Century: An Encyclopedia. ABC-CLIO. ISBN 978-1-85109-665-7. Archived from the original on 2023-10-28. Retrieved 2015-07-27.
- Helvoort, Ton van (2000). Arne Hessenbruch (ed.). Reader's Guide to the History of Science. Fitzroy Dearborn Publishing. ISBN 978-1-884964-29-9. Archived from the original on 2023-10-28. Retrieved 2015-07-27.
- Holmes, Frederic Lawrence (1987). Lavoisier and the Chemistry of Life: An Exploration of Scientific Creativity. University of Wisconsin Press. ISBN 978-0-299-09984-8. Archived from the original on 2023-10-28. Retrieved 2015-07-27.
- Horton, Derek, ed. (2013). Advances in Carbohydrate Chemistry and Biochemistry, Volume 70. Academic Press. ISBN 978-0-12-408112-3. Archived from the original on 2023-10-28. Retrieved 2015-07-27.
- Hunter, Graeme K. (2000). Vital Forces: The Discovery of the Molecular Basis of Life. Academic Press. ISBN 978-0-12-361811-5. Archived from the original on 2023-10-28. Retrieved 2020-06-05.
- Karp, Gerald (2009). Cell and Molecular Biology: Concepts and Experiments. John Wiley & Sons. ISBN 978-0-470-48337-4.
- Kauffman, George B.; Chooljian, Steven H. (2001). "Friedrich Wöhler (1800–1882), on the Bicentennial of His Birth". The Chemical Educator. 6 (2): 121–133. doi:10.1007/s00897010444a. S2CID 93425404.
- Kleinkauf, Horst; Döhren, Hans von; Jaenicke Lothar (1988). The Roots of Modern Biochemistry: Fritz Lippmann's Squiggle and its Consequences. Walter de Gruyter & Co. p. 116. ISBN 978-3-11-085245-5. Archived from the original on 2023-10-28. Retrieved 2015-07-27.
- Knowles, J.R. (1980). "Enzyme-Catalyzed Phosphoryl Transfer Reactions". Annual Review of Biochemistry. 49: 877–919. doi:10.1146/annurev.bi.49.070180.004305. PMID 6250450. S2CID 7452392.
- Metzler, David Everett; Metzler, Carol M. (2001). Biochemistry: The Chemical Reactions of Living Cells. Vol. 1. Academic Press. ISBN 978-0-12-492540-3.
- Miller G; Spoolman Scott (2012). Environmental Science – Biodiversity Is a Crucial Part of the Earth's Natural Capital. Cengage Learning. ISBN 978-1-133-70787-5. Archived from the original on 2023-04-13. Retrieved 2016-01-04.
- Nielsen, Forrest H. (1999). "Ultratrace minerals". In Maurice E. Shils; et al. (eds.). Modern Nutrition in Health and Disease. Baltimore: Williams & Wilkins. pp. 283–303. hdl:10113/46493.
- Peet, Alisa (2012). Marks, Allan; Lieberman Michael A. (eds.). Marks' Basic Medical Biochemistry (Lieberman, Marks's Basic Medical Biochemistry) (4th ed.). Lippincott Williams & Wilkins. ISBN 978-1-60831-572-7. Archived from the original on 2023-10-28. Retrieved 2020-06-05.
- Rayner-Canham, Marelene F.; Rayner-Canham, Marelene; Rayner-Canham, Geoffrey (2005). Women in Chemistry: Their Changing Roles from Alchemical Times to the Mid-Twentieth Century. Chemical Heritage Foundation. ISBN 978-0-941901-27-7.
- Rojas-Ruiz, Fernando A.; Vargas-Méndez, Leonor Y.; Kouznetsov, Vladimir V. (2011). "Challenges and Perspectives of Chemical Biology, a Successful Multidisciplinary Field of Natural Sciences". Molecules. 16 (3): 2672–2687. doi:10.3390/molecules16032672. PMC 6259834. PMID 21441869.
- Saenger, Wolfram (1984). Principles of Nucleic Acid Structure. New York: Springer-Verlag. ISBN 978-0-387-90762-8. Archived from the original on 2023-10-28. Retrieved 2020-06-05.
- Slabaugh, Michael R.; Seager, Spencer L. (2013). Organic and Biochemistry for Today (6th ed.). Pacific Grove: Brooks Cole. ISBN 978-1-133-60514-0. Archived from the original on 2023-10-28. Retrieved 2020-06-05.
- Sherwood, Lauralee; Klandorf, Hillar; Yancey, Paul H. (2012). Animal Physiology: From Genes to Organisms. Cengage Learning. ISBN 978-0-8400-6865-1.
- Stryer L, Berg JM, Tymoczko JL (2007). Biochemistry (6th ed.). San Francisco: W.H. Freeman. ISBN 978-0-7167-8724-2.
- Tropp, Burton E. (2012). Molecular Biology (4th ed.). Jones & Bartlett Learning. ISBN 978-1-4496-0091-4. Archived from the original on 2023-10-28. Retrieved 2020-06-05.
- UNICEF (2010). Facts for life (PDF) (4th ed.). New York: United Nations Children's Fund. ISBN 978-92-806-4466-1. Archived (PDF) from the original on 2022-10-09.
- Ulveling, Damien; Francastel, Claire; Hubé, Florent (2011). "When one is better than two: RNA with dual functions" (PDF). Biochimie. 93 (4): 633–644. doi:10.1016/j.biochi.2010.11.004. PMID 21111023. S2CID 22165949. Archived (PDF) from the original on 2022-10-09.
- Varki A, Cummings R, Esko J, Jessica F, Hart G, Marth J (1999). Essentials of glycobiology. Cold Spring Harbor Laboratory Press. ISBN 978-0-87969-560-6. Archived from the original on 2023-10-28. Retrieved 2020-06-05.
- Voet, D; Voet, JG (2005). Biochemistry (3rd ed.). Hoboken, NJ: John Wiley & Sons Inc. ISBN 978-0-471-19350-0. Archived from the original on September 11, 2007.
- Whiting, G.C (1970). "Sugars". In A.C. Hulme (ed.). The Biochemistry of Fruits and their Products. Vol. 1. London & New York: Academic Press. ISBN 978-0-12-361201-4.
- Ziesak, Anne-Katrin; Cram Hans-Robert (1999). Walter de Gruyter Publishers, 1749–1999. Walter de Gruyter & Co. ISBN 978-3-11-016741-2. Archived from the original on 2023-10-28. Retrieved 2015-07-27.
- Ashcroft, Steve. "Professor Sir Philip Randle; Researcher into metabolism: [1st Edition]". Independent. ProQuest 311080685.
Further reading
[edit]- Fruton, Joseph S. Proteins, Enzymes, Genes: The Interplay of Chemistry and Biology. Yale University Press: New Haven, 1999. ISBN 0-300-07608-8
- Keith Roberts, Martin Raff, Bruce Alberts, Peter Walter, Julian Lewis and Alexander Johnson, Molecular Biology of the Cell
- 4th Edition, Routledge, March, 2002, hardcover, 1616 pp. ISBN 0-8153-3218-1
- 3rd Edition, Garland, 1994, ISBN 0-8153-1620-8
- 2nd Edition, Garland, 1989, ISBN 0-8240-3695-6
- Kohler, Robert. From Medical Chemistry to Biochemistry: The Making of a Biomedical Discipline. Cambridge University Press, 1982.
- Maggio, Lauren A.; Willinsky, John M.; Steinberg, Ryan M.; Mietchen, Daniel; Wass, Joseph L.; Dong, Ting (2017). "Wikipedia as a gateway to biomedical research: The relative distribution and use of citations in the English Wikipedia". PLOS ONE. 12 (12) e0190046. Bibcode:2017PLoSO..1290046M. doi:10.1371/journal.pone.0190046. PMC 5739466. PMID 29267345.
External links
[edit]- "Biochemical Society".
- The Virtual Library of Biochemistry, Molecular Biology and Cell Biology
- Biochemistry, 5th ed. Full text of Berg, Tymoczko, and Stryer, courtesy of NCBI.
- SystemsX.ch – The Swiss Initiative in Systems Biology
- Full text of Biochemistry by Kevin and Indira, an introductory biochemistry textbook.
Biochemistry
View on GrokipediaHistory
Origins and Early Discoveries
The roots of biochemistry emerged from longstanding philosophical debates in the 18th and early 19th centuries between vitalism, which posited that living organisms were governed by a non-physical "vital force" distinct from ordinary chemical processes, and mechanism, which advocated that life could be explained through physical and chemical laws alone.[6] This tension shaped early efforts to study biological phenomena chemically, as vitalists argued that organic compounds could only arise in living systems, while mechanists sought to bridge the gap between inorganic and organic chemistry.[7] A pivotal moment came in 1828 when German chemist Friedrich Wöhler synthesized urea—an organic compound found in urine—from inorganic ammonium cyanate, demonstrating that complex biological molecules could be produced in a laboratory without vital intervention.[8] This experiment challenged vitalist doctrines by showing continuity between inorganic and organic realms, paving the way for viewing life as a series of chemical transformations, though its immediate impact on vitalism's decline was more symbolic than revolutionary.[8] Building on such insights, French chemist Antoine Lavoisier conducted groundbreaking respiration experiments in the 1770s, measuring oxygen consumption and carbon dioxide production in animals and humans, which framed breathing as a form of slow combustion akin to chemical oxidation.[9] These studies established metabolism as a quantifiable chemical process, linking physiological functions to elemental reactions and influencing later biochemical inquiries into energy transformation.[10] In the 1840s, German chemist Justus von Liebig advanced these ideas through his work in physiological or "animal" chemistry, analyzing the chemical composition of foods and bodily fluids to elucidate metabolic pathways in nutrition and respiration.[11] Liebig's experiments demonstrated that animal heat and work arose from the oxidation of carbon and hydrogen in foodstuffs, treating organisms as chemical engines and emphasizing the role of nitrogenous compounds in tissue repair.[12] Toward the late 19th century, Emil Fischer's structural elucidations of sugars like glucose and proteins, including peptide bond formations, provided foundational models for biomolecular architecture, revealing life's building blocks as intricate yet chemically analyzable entities.[13] A culminating discovery occurred in 1897 when Eduard Buchner extracted a yeast press-juice that fermented sugar into alcohol and carbon dioxide without intact cells, proving that enzymatic processes could operate extracorporeally and solidifying biochemistry's focus on isolated chemical mechanisms in biology.[14] These milestones collectively shifted perceptions, portraying vital processes as governed by chemistry rather than mystical forces.[15]Development of Key Concepts and Techniques
The elucidation of the citric acid cycle by Hans Krebs in 1937 marked a pivotal advancement in understanding cellular respiration, demonstrating how acetate is oxidized through a series of enzymatic reactions involving citric acid intermediates to generate energy. Krebs' work, building on earlier observations of carbohydrate metabolism, integrated organic chemistry with physiology and earned him the 1953 Nobel Prize in Physiology or Medicine, shared with Fritz Lipmann for discoveries on coenzyme A. This cycle became a cornerstone for subsequent metabolic pathway research, highlighting the interconnectedness of biochemical processes. In the 1940s and 1950s, Linus Pauling advanced protein structural biology by proposing the alpha-helix and beta-sheet configurations based on X-ray diffraction data and quantum mechanical principles, revolutionizing the understanding of polypeptide folding. Pauling's models emphasized hydrogen bonding's role in secondary structures, influencing later studies on protein function and earning recognition in his 1954 Nobel Prize in Chemistry for chemical bond research. Concurrently, James Watson and Francis Crick's 1953 double-helix model of DNA provided a structural basis for genetic information storage and replication, integrating biochemical and crystallographic evidence from Rosalind Franklin's work. Their discovery, published in Nature, laid the foundation for molecular biology and was honored with the 1962 Nobel Prize in Physiology or Medicine, shared with Maurice Wilkins. Technological innovations in the mid-20th century further propelled biochemical analysis. The development of gel electrophoresis in the 1950s, pioneered by Oliver Smithies for protein separation based on charge and size, enabled precise purification and characterization of biomolecules, earning Arne Tiselius the 1948 Nobel Prize in Chemistry for foundational electrophoretic methods. By the 1970s, nuclear magnetic resonance (NMR) spectroscopy emerged as a non-destructive tool for determining molecular structures in solution, with Kurt Wüthrich's applications to proteins in the 1980s resolving three-dimensional folds and contributing to his 2002 Nobel Prize in Chemistry. These techniques democratized structural biochemistry, allowing detailed studies of enzyme-substrate interactions and nucleic acid conformations. The 2010s introduced CRISPR-Cas9 as a transformative tool for biochemical manipulation, with Jennifer Doudna and Emmanuelle Charpentier's 2012 demonstration of RNA-guided DNA cleavage enabling precise gene editing and functional genomics studies. Their innovation, recognized with the 2020 Nobel Prize in Chemistry shared with Charpentier, facilitated biochemical investigations into gene regulation and protein expression. Nobel recognitions underscored these shifts: the 1975 Chemistry Prize to John Cornforth for his studies on the stereochemistry of enzyme-catalyzed reactions,[16] the 1980 Chemistry Prize to Paul Berg, Walter Gilbert, and Frederick Sanger for their contributions to the biochemistry of nucleic acids, including recombinant DNA and sequencing,[17] the 2023 Physiology or Medicine Prize to Katalin Karikó and Drew Weissman for discoveries concerning nucleoside base modifications that enabled the development of effective mRNA vaccines, and the 2024 Chemistry Prize to David Baker, Demis Hassabis, and John Jumper for computational protein design and protein structure prediction.[18] Post-2000, biochemistry evolved toward integrative approaches like systems biochemistry, which models network-level interactions using computational tools to predict cellular responses. Metabolomics, advanced through high-throughput mass spectrometry and NMR, profiles small-molecule metabolites to map dynamic pathways, as exemplified in global human metabolome projects initiated around 2007. A landmark in predictive modeling came with DeepMind's AlphaFold in 2020, achieving near-experimental accuracy in protein structure prediction via AI, transforming structural genomics and drug design. These developments, incorporating genomic data, have unified biochemistry with systems biology, enabling holistic views of cellular function.Chemical Foundations
Essential Elements and Atoms
Living organisms are primarily composed of a limited set of chemical elements, with six elements—oxygen (O), carbon (C), hydrogen (H), nitrogen (N), calcium (Ca), and phosphorus (P)—accounting for approximately 99% of the mass in the human body. The elements carbon (C), hydrogen (H), oxygen (O), nitrogen (N), phosphorus (P), and sulfur (S), collectively known as CHNOPS, form the foundational building blocks of biological molecules, enabling the complexity and functionality of life. Oxygen dominates by mass at approximately 65%, largely due to its prevalence in water, which constitutes 60-70% of body weight, while carbon makes up about 18%, serving as the structural backbone for organic compounds. Hydrogen and nitrogen follow at roughly 10% and 3%, respectively, contributing to water, organic structures, and key biomolecules like proteins and nucleic acids. Calcium, at approximately 1.5%, is essential for bone mineralization and cellular signaling. Phosphorus and sulfur are present in smaller amounts, at about 1% and 0.25%, yet play critical roles in energy transfer molecules such as ATP and in amino acids like cysteine and methionine. The roles of these elements are tightly linked to their chemical properties. Carbon's versatility stems from its ability to form four stable covalent bonds, arranged in a tetrahedral geometry that allows for diverse three-dimensional structures in biomolecules.[19] This bonding capacity, combined with moderate electronegativity (2.55 on the Pauling scale), enables carbon to create stable chains and rings essential for life's molecular diversity.[20] Hydrogen (electronegativity 2.20) readily forms nonpolar bonds with carbon but polar bonds with more electronegative atoms like oxygen (3.44) and nitrogen (3.04), facilitating hydrogen bonding crucial for molecular interactions.[21] Phosphorus (2.19) and sulfur (2.58) contribute to high-energy bonds and disulfide bridges, respectively, due to their ability to form multiple oxidation states and polar linkages.[22] Besides these major elements, trace elements constitute less than 1% of body mass but are indispensable for specific functions. For instance, iron (Fe), at under 0.01% of total mass, is central to hemoglobin, where it binds oxygen reversibly through redox changes between Fe²⁺ and Fe³⁺ states, enabling efficient transport in blood.[23] Magnesium (Mg), comprising about 0.05%, forms the core of chlorophyll in plants, coordinating with nitrogenous ligands to absorb light for photosynthesis.[24] These elements' low abundance belies their catalytic and structural importance, often as cofactors in enzymes. Isotopic variations of these elements also hold biological significance. Carbon-14 (¹⁴C), a radioactive isotope with a half-life of 5,730 years, is incorporated into biomolecules during life via atmospheric CO₂ fixation and decays post-mortem, allowing radiocarbon dating to determine the age of organic remains up to about 50,000 years old.[25] This technique has revolutionized biochemical studies of ancient ecosystems and molecular turnover rates, providing insights into evolutionary timelines without altering the primary elemental composition.[26]| Element | Approximate % by Mass in Human Body | Key Biological Role |
|---|---|---|
| Oxygen (O) | 65 | Component of water and organic molecules; enables respiration |
| Carbon (C) | 18 | Backbone of organic structures |
| Hydrogen (H) | 10 | In water and C-H bonds for energy storage |
| Nitrogen (N) | 3 | In amino acids and nucleic acids |
| Calcium (Ca) | 1.5 | Structural component in bones and teeth; cellular signaling |
| Phosphorus (P) | 1 | In ATP, DNA, and phospholipids |
| Sulfur (S) | 0.25 | In cysteine, methionine, and coenzymes |
| Iron (Fe) | <0.01 | Oxygen transport in hemoglobin |
| Magnesium (Mg) | 0.05 | Central ion in chlorophyll; enzyme cofactor |






