Recent from talks
Contribute something
Nothing was collected or created yet.
Virus
View on Wikipedia
| Virus | |
|---|---|

| |
| SARS-CoV-2, a member of the subfamily Orthocoronavirinae | |
| Virus classification | |
| (unranked): | Virus |
| Realms | |
A virus is a submicroscopic infectious agent that replicates only inside the living cells of an organism.[1] Viruses infect all life forms, from animals and plants to microorganisms, including bacteria and archaea.[2][3] Viruses are found in almost every ecosystem on Earth and are the most numerous type of biological entity.[4][5] Since Dmitri Ivanovsky's 1892 article describing a non-bacterial pathogen infecting tobacco plants and the discovery of the tobacco mosaic virus by Martinus Beijerinck in 1898,[6]: 4 more than 16,000 of the millions of virus species have been described in detail.[7][8] The study of viruses is known as virology, a subspeciality of microbiology.
When infected, a host cell is often forced to rapidly produce thousands of copies of the original virus. When not inside an infected cell or in the process of infecting a cell, viruses exist in the form of independent viral particles, or virions, consisting of (i) genetic material, i.e., long molecules of DNA or RNA that encode the structure of the proteins by which the virus acts; (ii) a protein coat, the capsid, which surrounds and protects the genetic material; and in some cases (iii) an outside envelope of lipids. The shapes of these virus particles range from simple helical and icosahedral forms to more complex structures. Most virus species have virions too small to be seen with an optical microscope and are one-hundredth the size of most bacteria.
The origins of viruses in the evolutionary history of life are still unclear. Some viruses may have evolved from plasmids, which are pieces of DNA that can move between cells. Other viruses may have evolved from bacteria. In evolution, viruses are an important means of horizontal gene transfer, which increases genetic diversity in a way analogous to sexual reproduction.[9] Viruses are considered by some biologists to be a life form, because they carry genetic material, reproduce, and evolve through natural selection, although they lack some key characteristics, such as cell structure, that are generally considered necessary criteria for defining life. Because they possess some but not all such qualities, viruses have been described as "organisms at the edge of life"[10] and as replicators.[11]
Viruses spread in many ways. One transmission pathway is through disease-bearing organisms known as vectors: for example, viruses are often transmitted from plant to plant by insects that feed on plant sap, such as aphids; and viruses in animals can be carried by blood-sucking insects. Many viruses spread in the air by coughing and sneezing, including influenza viruses, SARS-CoV-2, chickenpox, smallpox, and measles. Norovirus and rotavirus, common causes of viral gastroenteritis, are transmitted by the faecal–oral route, passed by hand-to-mouth contact or in food or water. The infectious dose of norovirus required to produce infection in humans is fewer than 100 particles.[12] HIV is one of several viruses transmitted through sexual contact and by exposure to infected blood. The variety of host cells that a virus can infect is called its host range: this is narrow for viruses specialized to infect only a few species, or broad for viruses capable of infecting many.[13]: 123–124
Viral infections in animals provoke an immune response that usually eliminates the infecting virus. Immune responses can also be produced by vaccines, which confer an artificially acquired immunity to the specific viral infection. Some viruses, including those that cause HIV/AIDS, HPV infection, and viral hepatitis, evade these immune responses and result in chronic infections. Several classes of antiviral drugs have been developed.
Etymology
[edit]The English word "virus" comes from the Latin word vīrus, which refers to poison and other noxious liquids. Vīrus comes from the same Indo-European root as Sanskrit viṣa, Avestan vīša, and Ancient Greek ἰός (iós), which all mean "poison". The first attested use of "virus" in English appeared in 1398 in John Trevisa's translation of Bartholomeus Anglicus's De Proprietatibus Rerum.[14][15] Virulent, from Latin virulentus ('poisonous'), dates to c. 1400.[16][17] A meaning of 'agent that causes infectious disease' is first recorded in 1728,[15] long before the discovery of viruses by Dmitri Ivanovsky in 1892. The English plural is viruses (sometimes also vira),[18] whereas the Latin word is a mass noun, which has no classically attested plural (vīra is used in Neo-Latin[19]). The adjective viral dates to 1948.[20] The term virion (plural virions), which dates from 1959,[21] is also used to refer to a single viral particle that is released from the cell and is capable of infecting other cells of the same type.[22]
Origins
[edit]Viruses are found wherever there is life and have probably existed since living cells first evolved.[23] The origin of viruses is unclear because they do not form fossils, so molecular techniques are used to infer how they arose.[24] In addition, viral genetic material occasionally integrates into the germline of the host organisms, by which they can be passed on vertically to the offspring of the host for many generations. This provides an invaluable source of information for paleovirologists to trace back ancient viruses that existed as far back as millions of years ago.
There are three main hypotheses that aim to explain the origins of viruses:[25]
- Regressive hypothesis
- Viruses may have once been small cells that parasitised larger cells. Over time, genes not required by their parasitism were lost. The bacteria rickettsia and chlamydia are living cells that, like viruses, can reproduce only inside host cells. They lend support to this hypothesis, as their dependence on parasitism is likely to have caused the loss of genes that enabled them to survive outside a cell. This is also called the "degeneracy hypothesis",[6]: 16 [26]: 11 or "reduction hypothesis".[27]: 24
- Cellular origin hypothesis
- Some viruses may have evolved from bits of DNA or RNA that "escaped" from the genes of a larger organism. The escaped DNA could have come from plasmids (pieces of naked DNA that can move between cells) or transposons (molecules of DNA that replicate and move around to different positions within the genes of the cell).[13]: 810 Once called "jumping genes", transposons are examples of mobile genetic elements and could be the origin of some viruses. They were discovered in maize by Barbara McClintock in 1950.[28] This is sometimes called the "vagrancy hypothesis",[6]: 16 [26]: 11–12 or the "escape hypothesis".[27]: 24
- Co-evolution hypothesis
- This is also called the "virus-first hypothesis"[27]: 24 and proposes that viruses may have evolved from complex molecules of protein and nucleic acid at the same time that cells first appeared on Earth and would have been dependent on cellular life for billions of years. Viroids are molecules of RNA that are not classified as viruses because they lack a protein coat. They have characteristics that are common to several viruses and are often called subviral agents.[6]: 55 Viroids are significant pathogens of plants.[13]: 791 They do not code for proteins but interact with the host cell and use the host machinery for their replication.[29] The hepatitis delta virus of humans has an RNA genome similar to viroids but has a protein coat derived from hepatitis B virus and cannot produce one of its own. It is, therefore, a defective virus. Although hepatitis delta virus genome may replicate independently once inside a host cell, it requires the help of hepatitis B virus to provide a protein coat so that it can be transmitted to new cells.[13]: 460 In similar manner, the sputnik virophage is dependent on mimivirus, which infects the protozoan Acanthamoeba castellanii.[30] These viruses, which are dependent on the presence of other virus species in the host cell, are called "satellites" and may represent evolutionary intermediates of viroids and viruses.[26]: 777 [6]: 55–57
In the past, there were problems with all of these hypotheses: the regressive hypothesis did not explain why even the smallest of cellular parasites do not resemble viruses in any way. The escape hypothesis did not explain the complex capsids and other structures on virus particles. The virus-first hypothesis contravened the definition of viruses in that they require host cells.[27]: 24 Viruses are now recognised as ancient and as having origins that pre-date the divergence of life into the three domains.[27]: 28 This discovery has led modern virologists to reconsider and re-evaluate these three classical hypotheses.[27]: 28
The evidence for an ancestral world of RNA cells[27]: 26 and computer analysis of viral and host DNA sequences give a better understanding of the evolutionary relationships between different viruses and may help identify the ancestors of modern viruses. To date, such analyses have not proved which of these hypotheses is correct.[27]: 26 It seems unlikely that all currently known viruses have a common ancestor, and viruses have probably arisen numerous times in the past by one or more mechanisms.[31]
Microbiology
[edit]Discovery
[edit]The first evidence of the existence of viruses came from experiments with filters that had pores small enough to retain bacteria. In 1892, Dmitri Ivanovsky used one of these filters to show that sap from a diseased tobacco plant remained infectious to healthy tobacco plants despite having been filtered. Martinus Beijerinck called the filtered, infectious substance a "virus" and this discovery is considered to be the beginning of virology. The subsequent discovery and partial characterization of bacteriophages by Frederick Twort and Félix d'Herelle further catalyzed the field, and by the early 20th century many viruses had been discovered. In 1926, Thomas Milton Rivers defined viruses as obligate parasites. Viruses were demonstrated to be particles, rather than a fluid, by Wendell Meredith Stanley, and the invention of the electron microscope in 1931 allowed their complex structures to be visualised.[32]
Life properties
[edit]Scientific opinions differ on whether viruses are a form of life or organic structures that interact with living organisms.[11] They have been described as "organisms at the edge of life",[10] since they resemble organisms in that they possess genes, evolve by natural selection,[33] and reproduce by creating multiple copies of themselves through self-assembly. Although they have genes, they do not have a cellular structure, which is often seen as the basic unit of life. Viruses do not have their own metabolism and require a host cell to make new products. They therefore cannot naturally reproduce outside a host cell[34]—although some bacteria such as rickettsia and chlamydia are considered living organisms despite the same limitation.[35][36] Accepted forms of life use cell division to reproduce, whereas viruses spontaneously assemble within cells. They differ from autonomous growth of crystals as they inherit genetic mutations while being subject to natural selection. Virus self-assembly within host cells has implications for the study of the origin of life, as it lends further credence to the hypothesis that life could have started as self-assembling organic molecules.[2] The virocell model first proposed by Patrick Forterre considers the infected cell to be the "living form" of viruses and that virus particles (virions) are analogous to spores.[37] Although the living versus non-living debate continues, the virocell model has gained some acceptance.[38]
Structure
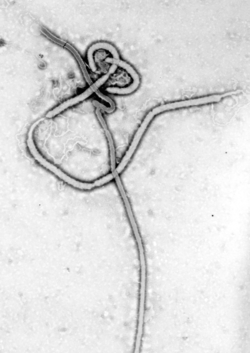
[edit]Viruses display a wide diversity of sizes and shapes, called 'morphologies'. In general, viruses are much smaller than bacteria and more than a thousand bacteriophage viruses would fit inside an Escherichia coli bacterium's cell.[39]: 98 Many viruses that have been studied are spherical and have a diameter between 20 and 300 nanometres. Some filoviruses, which are filaments, have a total length of up to 1400 nm; their diameters are only about 80 nm.[26]: 33–55 Most viruses cannot be seen with an optical microscope, so scanning and transmission electron microscopes are used to visualise them.[26]: 33–37 To increase the contrast between viruses and the background, electron-dense "stains" are used. These are solutions of salts of heavy metals, such as tungsten, that scatter the electrons from regions covered with the stain. When virions are coated with stain (positive staining), fine detail is obscured. Negative staining overcomes this problem by staining the background only.[40]
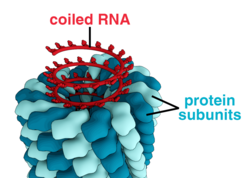
A complete virus particle, known as a virion, consists of nucleic acid surrounded by a protective coat of protein called a capsid. These are formed from protein subunits called capsomeres.[26]: 40 Viruses can have a lipid "envelope" derived from the host cell membrane. The capsid is made from proteins encoded by the viral genome and its shape serves as the basis for morphological distinction.[41][42] Virally-coded protein subunits will self-assemble to form a capsid, in general requiring the presence of the virus genome. Complex viruses code for proteins that assist in the construction of their capsid. Proteins associated with nucleic acid are known as nucleoproteins, and the association of viral capsid proteins with viral nucleic acid is called a nucleocapsid. The capsid and entire virus structure can be mechanically (physically) probed through atomic force microscopy.[43][44] In general, there are five main morphological virus types:
- Helical
- These viruses are composed of a single type of capsomere stacked around a central axis to form a helical structure, which may have a central cavity, or tube. This arrangement results in virions which can be short and highly rigid rods, or long and very flexible filaments. The genetic material (typically single-stranded RNA, but single-stranded DNA in some cases) is bound into the protein helix by interactions between the negatively charged nucleic acid and positive charges on the protein. Overall, the length of a helical capsid is related to the length of the nucleic acid contained within it, and the diameter is dependent on the size and arrangement of capsomeres. The well-studied tobacco mosaic virus[26]: 37 and inovirus[45] are examples of helical viruses.
- Icosahedral
- Most animal viruses are icosahedral or near-spherical with chiral icosahedral symmetry. A regular icosahedron is the optimum way of forming a closed shell from identical subunits. The minimum number of capsomeres required for each triangular face is 3, which gives 60 for the icosahedron. Many viruses, such as rotavirus, have more than 60 capsomers and appear spherical but they retain this symmetry. To achieve this, the capsomeres at the apices are surrounded by five other capsomeres and are called pentons. Capsomeres on the triangular faces are surrounded by six others and are called hexons.[26]: 40, 42 Hexons are in essence flat and pentons, which form the 12 vertices, are curved. The same protein may act as the subunit of both the pentamers and hexamers or they may be composed of different proteins.[46]
- Prolate
- This is an icosahedron elongated along the fivefold axis and is a common arrangement of the heads of bacteriophages. This structure is composed of a cylinder with a cap at either end.[47]
- Enveloped
- Some species of virus envelop themselves in a modified form of one of the cell membranes, either the outer membrane surrounding an infected host cell or internal membranes such as a nuclear membrane or endoplasmic reticulum, thus gaining an outer lipid bilayer known as a viral envelope. This membrane is studded with proteins coded for by the viral genome and host genome; the lipid membrane itself and any carbohydrates present originate entirely from the host. Influenza virus, HIV (which causes AIDS), and severe acute respiratory syndrome coronavirus 2 (which causes COVID-19)[48] use this strategy. Most enveloped viruses are dependent on the envelope for their infectivity.[26]: 42–43
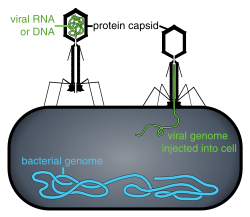
- Complex
- These viruses possess a capsid that is neither purely helical nor purely icosahedral, and that may possess extra structures such as protein tails or a complex outer wall. Some bacteriophages, such as Enterobacteria phage T4, have a complex structure consisting of an icosahedral head bound to a helical tail, which may have a hexagonal base plate with protruding protein tail fibres. This tail structure acts like a molecular syringe, attaching to the bacterial host and then injecting the viral genome into the cell.[49]
The poxviruses are large, complex viruses that have an unusual morphology. The viral genome is associated with proteins within a central disc structure known as a nucleoid. The nucleoid is surrounded by a membrane and two lateral bodies of unknown function. The virus has an outer envelope with a thick layer of protein studded over its surface. The whole virion is slightly pleomorphic, ranging from ovoid to brick-shaped.[50]
Giant viruses
[edit]Mimivirus is one of the largest characterised viruses, with a capsid diameter of 400 nm. Protein filaments measuring 100 nm project from the surface. The capsid appears hexagonal under an electron microscope, therefore the capsid is probably icosahedral.[51] In 2011, researchers discovered the largest then known virus in samples of water collected from the ocean floor off the coast of Las Cruces, Chile. Provisionally named Megavirus chilensis, it can be seen with a basic optical microscope.[52] In 2013, the Pandoravirus genus was discovered in Chile and Australia, and has genomes about twice as large as Megavirus and Mimivirus.[53] All giant viruses have dsDNA genomes and they are classified into several families: Mimiviridae, Pithoviridae, Pandoraviridae, Phycodnaviridae, and the Mollivirus genus.[54]
Some viruses that infect Archaea have complex structures unrelated to any other form of virus, with a wide variety of unusual shapes, ranging from spindle-shaped structures to viruses that resemble hooked rods, teardrops or even bottles. Other archaeal viruses resemble the tailed bacteriophages, and can have multiple tail structures.[55]
Genome
[edit]| Property | Parameters |
|---|---|
| Nucleic acid |
|
| Shape |
|
| Strandedness |
|
| Sense |
|
An enormous variety of genomic structures can be seen among viral species; as a group, they contain more structural genomic diversity than plants, animals, archaea, or bacteria. There are millions of different types of viruses,[8] although fewer than 7,000 types have been described in detail.[6]: 49 As of January 2021, the NCBI Virus genome database has more than 193,000 complete genome sequences,[56] but there are doubtlessly many more to be discovered.[57][58]
A virus has either a DNA or an RNA genome and is called a DNA virus or an RNA virus, respectively. Some RNA viruses, for example retroviruses, have a stage in their replication cycle where the genome is encoded in DNA.[59] Most viruses have RNA genomes. Plant viruses tend to have single-stranded RNA genomes and bacteriophages tend to have double-stranded DNA genomes.[26]: 96–99
Viral genomes are circular, as in the polyomaviruses, or linear, as in the adenoviruses. The type of nucleic acid is irrelevant to the shape of the genome. Among RNA viruses and certain DNA viruses, the genome is often divided into separate parts, in which case it is called segmented. For RNA viruses, each segment often codes for only one protein and they are usually found together in one capsid. All segments are not required to be in the same virion for the virus to be infectious, as demonstrated by brome mosaic virus and several other plant viruses.[26]: 33–35
A viral genome, irrespective of nucleic acid type, is almost always either single-stranded (ss) or double-stranded (ds). Single-stranded genomes consist of an unpaired nucleic acid, analogous to one-half of a ladder split down the middle. Double-stranded genomes consist of two complementary paired nucleic acids, analogous to a ladder. The virus particles of some virus families, such as those belonging to the Hepadnaviridae, contain a genome that is partially double-stranded and partially single-stranded.[26]: 96–99
For most viruses with RNA genomes and some with single-stranded DNA (ssDNA) genomes, the single strands are said to be either positive-sense (called the 'plus-strand') or negative-sense (called the 'minus-strand'), depending on if they are complementary to the viral messenger RNA (mRNA). Positive-sense viral RNA is in the same sense as viral mRNA and thus at least a part of it can be immediately translated by the host cell. Negative-sense viral RNA is complementary to mRNA and thus must be converted to positive-sense RNA by an RNA-dependent RNA polymerase before translation. DNA nomenclature for viruses with genomic ssDNA is similar to RNA nomenclature, in that positive-strand viral ssDNA is identical in sequence to the viral mRNA and is thus a coding strand, while negative-sense viral ssDNA is complementary to the viral mRNA and is thus a template strand.[26]: 96–99 Several types of ssDNA and ssRNA viruses have genomes that are ambisense in that transcription can occur off both strands in a double-stranded replicative intermediate. Examples include geminiviruses, which are ssDNA plant viruses and arenaviruses, which are ssRNA viruses of animals.[60]
Genome size
[edit]Genome size varies greatly between species. The smallest—the ssDNA circoviruses, family Circoviridae—code for only two proteins and have a genome size of only two kilobases;[61] the largest—the pandoraviruses—have genome sizes of around two megabases which code for about 2500 proteins.[53] Virus genes rarely have introns and often are arranged in the genome so that they overlap.[62]
In general, RNA viruses have smaller genome sizes than DNA viruses because of a higher error-rate when replicating, and have a maximum upper size limit.[24] Beyond this, errors when replicating render the virus useless or uncompetitive. To compensate, RNA viruses often have segmented genomes—the genome is split into smaller molecules—thus reducing the chance that an error in a single-component genome will incapacitate the entire genome. In contrast, DNA viruses generally have larger genomes because of the high fidelity of their replication enzymes.[63] Single-strand DNA viruses are an exception to this rule, as mutation rates for these genomes can approach the extreme of the ssRNA virus case.[64]
Genetic mutation and recombination
[edit]
Viruses undergo genetic change by several mechanisms. These include a process called antigenic drift where individual bases in the DNA or RNA mutate to other bases. Most of these point mutations are "silent"—they do not change the protein that the gene encodes—but others can confer evolutionary advantages such as resistance to antiviral drugs.[65][66] Antigenic shift occurs when there is a major change in the genome of the virus. This can be a result of recombination or reassortment. The Influenza A virus is highly prone to reassortment; occasionally this has resulted in novel strains which have caused pandemics.[67] RNA viruses often exist as quasispecies or swarms of viruses of the same species but with slightly different genome nucleoside sequences. Such quasispecies are a prime target for natural selection.[68]
Segmented genomes confer evolutionary advantages; different strains of a virus with a segmented genome can shuffle and combine genes and produce progeny viruses (or offspring) that have unique characteristics. This is called reassortment or 'viral sex'.[69]
Genetic recombination is a process by which a strand of DNA (or RNA) is broken and then joined to the end of a different DNA (or RNA) molecule. This can occur when viruses infect cells simultaneously and studies of viral evolution have shown that recombination has been rampant in the species studied.[70] Recombination is common to both RNA and DNA viruses.[71][72]
Coronaviruses have a single-strand positive-sense RNA genome. Replication of the genome is catalyzed by an RNA-dependent RNA polymerase. The mechanism of recombination used by coronaviruses likely involves template switching by the polymerase during genome replication.[73] This process appears to be an adaptation for coping with genome damage.[74]
Replication cycle
[edit]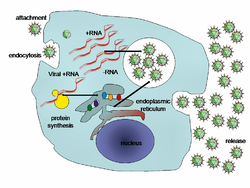
Viral populations do not grow through cell division, because they are acellular. Instead, they use the machinery and metabolism of a host cell to produce multiple copies of themselves, and they assemble in the cell.[75] When infected, the host cell is forced to rapidly produce thousands of copies of the original virus.[76]
Their life cycle differs greatly between species, but there are six basic stages in their life cycle:[26]: 75–91
Attachment is a specific binding between viral capsid proteins and specific receptors on the host cellular surface. This specificity determines the host range and type of host cell of a virus. For example, HIV infects a limited range of human leucocytes. This is because its surface protein, gp120, specifically interacts with the CD4 molecule—a chemokine receptor—which is most commonly found on the surface of CD4+ T-Cells. This mechanism has evolved to favour those viruses that infect only cells in which they are capable of replication. Attachment to the receptor can induce the viral envelope protein to undergo changes that result in the fusion of viral and cellular membranes, or changes of non-enveloped virus surface proteins that allow the virus to enter.[77]
Penetration or viral entry follows attachment: Virions enter the host cell through receptor-mediated endocytosis or membrane fusion. The infection of plant and fungal cells is different from that of animal cells. Plants have a rigid cell wall made of cellulose, and fungi one of chitin, so most viruses can get inside these cells only after trauma to the cell wall.[6]: 70 Nearly all plant viruses (such as tobacco mosaic virus) can also move directly from cell to cell, in the form of single-stranded nucleoprotein complexes, through pores called plasmodesmata.[78] Bacteria, like plants, have strong cell walls that a virus must breach to infect the cell. Given that bacterial cell walls are much thinner than plant cell walls due to their much smaller size, some viruses have evolved mechanisms that inject their genome into the bacterial cell across the cell wall, while the viral capsid remains outside.[6]: 71
Uncoating is a process in which the viral capsid is removed: This may be by degradation by viral enzymes or host enzymes or by simple dissociation; the end-result is the releasing of the viral genomic nucleic acid.[79]
Replication of viruses involves primarily multiplication of the genome. Replication involves the synthesis of viral messenger RNA (mRNA) from "early" genes (with exceptions for positive-sense RNA viruses), viral protein synthesis, possible assembly of viral proteins, then viral genome replication mediated by early or regulatory protein expression. This may be followed, for complex viruses with larger genomes, by one or more further rounds of mRNA synthesis: "late" gene expression is, in general, of structural or virion proteins.[80]
Assembly – Following the structure-mediated self-assembly of the virus particles, some modification of the proteins often occurs. In viruses such as HIV, this modification (sometimes called maturation) occurs after the virus has been released from the host cell.[81]
Release – Viruses can be released from the host cell by lysis, a process that kills the cell by bursting its membrane and cell wall if present: this is a feature of many bacterial and some animal viruses. Some viruses undergo a lysogenic cycle where the viral genome is incorporated by genetic recombination into a specific place in the host's chromosome. The viral genome is then known as a "provirus" or, in the case of bacteriophages a "prophage".[13]: 836 Whenever the host divides, the viral genome is also replicated. The viral genome is mostly silent within the host. At some point, the provirus or prophage may give rise to the active virus, which may lyse the host cells.[6]: 243–259 Enveloped viruses (e.g., HIV) typically are released from the host cell by budding. During this process, the virus acquires its envelope, which is a modified piece of the host's plasma or other, internal membrane.[6]: 185–187
Genome replication
[edit]The genetic material within virus particles, and the method by which the material is replicated, varies considerably between different types of viruses.
- DNA viruses
- The genome replication of most DNA viruses takes place in the cell's nucleus. If the cell has the appropriate receptor on its surface, these viruses enter the cell either by direct fusion with the cell membrane (e.g., herpesviruses) or—more usually—by receptor-mediated endocytosis. Most DNA viruses are entirely dependent on the host cell's DNA and RNA synthesising machinery and RNA processing machinery. Viruses with larger genomes may encode much of this machinery themselves. In eukaryotes, the viral genome must cross the cell's nuclear membrane to access this machinery, while in bacteria it need only enter the cell.[13]: 118 [26]: 78
- RNA viruses
- Replication of RNA viruses usually takes place in the cytoplasm. RNA viruses can be placed into four different groups depending on their modes of replication. The polarity (whether or not it can be used directly by ribosomes to make proteins) of single-stranded RNA viruses largely determines the replicative mechanism; the other major criterion is whether the genetic material is single-stranded or double-stranded. All RNA viruses use their own RNA replicase enzymes to create copies of their genomes.[26]: 79
- Reverse transcribing viruses
- Reverse transcribing viruses have ssRNA (Retroviridae, Metaviridae, Pseudoviridae) or dsDNA (Caulimoviridae, and Hepadnaviridae) in their particles. Reverse transcribing viruses with RNA genomes (retroviruses) use a DNA intermediate to replicate, whereas those with DNA genomes (pararetroviruses) use an RNA intermediate during genome replication. Both types use a reverse transcriptase, or RNA-dependent DNA polymerase enzyme, to carry out the nucleic acid conversion. Retroviruses integrate the DNA produced by reverse transcription into the host genome as a provirus as a part of the replication process; pararetroviruses do not, although integrated genome copies of especially plant pararetroviruses can give rise to infectious virus.[82] They are susceptible to antiviral drugs that inhibit the reverse transcriptase enzyme, e.g. zidovudine and lamivudine. An example of the first type is HIV, which is a retrovirus. Examples of the second type are the Hepadnaviridae, which includes Hepatitis B virus.[26]: 88–89
Cytopathic effects on the host cell
[edit]The range of structural and biochemical effects that viruses have on the host cell is extensive.[26]: 115–146 These are called 'cytopathic effects'.[26]: 115 Most virus infections eventually result in the death of the host cell. The causes of death include cell lysis, alterations to the cell's surface membrane and apoptosis.[83] Often cell death is caused by cessation of its normal activities because of suppression by virus-specific proteins, not all of which are components of the virus particle.[84] The distinction between cytopathic and harmless is gradual. Some viruses, such as Epstein–Barr virus, can cause cells to proliferate without causing malignancy,[85] while others, such as papillomaviruses, are established causes of cancer.[86]
Dormant and latent infections
[edit]Some viruses cause no apparent changes to the infected cell. Cells in which the virus is latent and inactive show few signs of infection and often function normally.[87] This causes persistent infections and the virus is often dormant for many months or years. This is often the case with herpes viruses.[88][89]
Host range
[edit]Viruses are by far the most abundant biological entities on Earth and they outnumber all the others put together.[90] They infect all types of cellular life including animals, plants, bacteria and fungi.[6]: 49 Different types of viruses can infect only a limited range of hosts and many are species-specific. Some, such as smallpox virus for example, can infect only one species—in this case humans,[13]: 643 and are said to have a narrow host range. Other viruses, such as rabies virus, can infect different species of mammals and are said to have a broad range.[13]: 631 The viruses that infect plants are harmless to animals, and most viruses that infect other animals are harmless to humans.[6]: 272 The host range of some bacteriophages is limited to a single strain of bacteria and they can be used to trace the source of outbreaks of infections by a method called phage typing.[91] The complete set of viruses in an organism or habitat is called the virome; for example, all human viruses constitute the human virome.[92]
Novel viruses
[edit]A novel virus is one that has not previously been recorded. It can be a virus that is isolated from its natural reservoir or isolated as the result of spread to an animal or human host where the virus had not been identified before. It can be an emergent virus, one that represents a new virus, but it can also be an extant virus that has not been previously identified.[93] The SARS-CoV-2 coronavirus that caused the COVID-19 pandemic is an example of a novel virus.[94]
Classification
[edit]Classification seeks to describe the diversity of viruses by naming and grouping them on the basis of similarities. In 1962, André Lwoff, Robert Horne, and Paul Tournier were the first to develop a means of virus classification, based on the Linnaean hierarchical system.[95] This system based classification on phylum, class, order, family, genus, and species. Viruses were grouped according to their shared properties (not those of their hosts) and the type of nucleic acid forming their genomes.[96] In 1966, the International Committee on Taxonomy of Viruses (ICTV) was formed. The system proposed by Lwoff, Horne and Tournier was initially not accepted by the ICTV because the small genome size of viruses and their high rate of mutation made it difficult to determine their ancestry beyond order. As such, the Baltimore classification system has come to be used to supplement the more traditional hierarchy.[97] Starting in 2018, the ICTV began to acknowledge deeper evolutionary relationships between viruses that have been discovered over time and adopted a 15-rank classification system ranging from realm to species.[98] Additionally, some species within the same genus are grouped into a genogroup.[99][100]
ICTV classification
[edit]The ICTV developed the current classification system and wrote guidelines that put a greater weight on certain virus properties to maintain family uniformity. A unified taxonomy (a universal system for classifying viruses) has been established.[101] Only a small part of the total diversity of viruses has been studied.[102] As of 2024, 7 realms, 11 kingdoms, 22 phyla, 4 subphyla, 49 classes, 93 orders, 12 suborders, 368 families, 213 subfamilies, 3,769 genera, 86 subgenera, and 16,215 species of viruses have been defined by the ICTV.[7]
The general taxonomic structure of taxon ranges and the suffixes used in taxonomic names are shown hereafter. As of 2022, the ranks of subrealm, subkingdom, and subclass are unused, whereas all other ranks are in use.[7]
- Realm (-viria)
Baltimore classification
[edit]
The Nobel Prize-winning biologist David Baltimore devised the Baltimore classification system.[103][104] The ICTV classification system is used in conjunction with the Baltimore classification system in modern virus classification.[105][106][107]
The Baltimore classification of viruses is based on the mechanism of mRNA production. Viruses must generate mRNAs from their genomes to produce proteins and replicate themselves, but different mechanisms are used to achieve this in each virus family. Viral genomes may be single-stranded (ss) or double-stranded (ds), RNA or DNA, and may or may not use reverse transcriptase (RT). In addition, ssRNA viruses may be either sense (+) or antisense (−). This classification places viruses into seven groups:
- I: dsDNA viruses (e.g. Adenoviruses, Herpesviruses, Poxviruses)
- II: ssDNA viruses (+ strand or "sense") DNA (e.g. Parvoviruses)
- III: dsRNA viruses (e.g. Reoviruses)
- IV:(+)ssRNA viruses (+ strand or sense) RNA (e.g. Coronaviruses, Picornaviruses, Togaviruses)
- V: (−)ssRNA viruses (− strand or antisense) RNA (e.g. Orthomyxoviruses, Rhabdoviruses)
- VI: ssRNA-RT viruses (+ strand or sense) RNA with DNA intermediate in life-cycle (e.g. Retroviruses)
- VII: dsDNA-RT viruses DNA with RNA intermediate in life-cycle (e.g. Hepadnaviruses)
Role in human disease
[edit]
Examples of common human diseases caused by viruses include the common cold, influenza, chickenpox, and cold sores. Many serious diseases such as rabies, Ebola virus disease, AIDS (HIV), avian influenza, and SARS are caused by viruses. The relative ability of viruses to cause disease is described in terms of virulence. Other diseases are under investigation to discover if they have a virus as the causative agent, such as the possible connection between human herpesvirus 6 (HHV6) and neurological diseases such as multiple sclerosis and chronic fatigue syndrome.[109] There is controversy over whether the bornavirus, previously thought to cause neurological diseases in horses, could be responsible for psychiatric illnesses in humans.[110]
Viruses have different mechanisms by which they produce disease in an organism, which depends largely on the viral species. Mechanisms at the cellular level primarily include cell lysis, the breaking open and subsequent death of the cell. In multicellular organisms, if enough cells die, the whole organism will start to suffer the effects. Although viruses cause disruption of healthy homeostasis, resulting in disease, they may exist relatively harmlessly within an organism. An example would include the ability of the herpes simplex virus, which causes cold sores, to remain in a dormant state within the human body. This is called latency[111] and is a characteristic of the herpes viruses, including Epstein–Barr virus, which causes glandular fever, and varicella zoster virus, which causes chickenpox and shingles. Most people have been infected with at least one of these types of herpes virus.[112] These latent viruses might sometimes be beneficial, as the presence of the virus can increase immunity against bacterial pathogens, such as Yersinia pestis.[113]
Some viruses can cause lifelong or chronic infections, where the viruses continue to replicate in the body despite the host's defence mechanisms.[114] This is common in hepatitis B virus and hepatitis C virus infections. People chronically infected are known as carriers, as they serve as reservoirs of infectious virus.[115] In populations with a high proportion of carriers, the disease is said to be endemic.[116]
Epidemiology
[edit]Viral epidemiology is the branch of medical science that deals with the transmission and control of virus infections in humans. Transmission of viruses can be vertical, which means from mother to child, or horizontal, which means from person to person. Examples of vertical transmission include hepatitis B virus and HIV, where the baby is born already infected with the virus.[117] Another, more rare, example is the varicella zoster virus, which, although causing relatively mild infections in children and adults, can be fatal to the foetus and newborn baby.[118]
Horizontal transmission is the most common mechanism of spread of viruses in populations.[119] Horizontal transmission can occur when body fluids are exchanged during sexual activity, by exchange of saliva or when contaminated food or water is ingested. It can also occur when aerosols containing viruses are inhaled or by insect vectors such as when infected mosquitoes penetrate the skin of a host.[119] Most types of viruses are restricted to just one or two of these mechanisms and they are referred to as "respiratory viruses" or "enteric viruses" and so forth. The rate or speed of transmission of viral infections depends on factors that include population density, the number of susceptible individuals, (i.e., those not immune),[120] the quality of healthcare and the weather.[121]
Epidemiology is used to break the chain of infection in populations during outbreaks of viral diseases.[13]: 264 Control measures are used that are based on knowledge of how the virus is transmitted. It is important to find the source, or sources, of the outbreak and to identify the virus. Once the virus has been identified, the chain of transmission can sometimes be broken by vaccines. When vaccines are not available, sanitation and disinfection can be effective. Often, infected people are isolated from the rest of the community, and those that have been exposed to the virus are placed in quarantine.[13]: 894 To control the outbreak of foot-and-mouth disease in cattle in Britain in 2001, thousands of cattle were slaughtered.[122] Most viral infections of humans and other animals have incubation periods during which the infection causes no signs or symptoms.[13]: 170 Incubation periods for viral diseases range from a few days to weeks, but are known for most infections.[13]: 170–172 Somewhat overlapping, but mainly following the incubation period, there is a period of communicability—a time when an infected individual or animal is contagious and can infect another person or animal.[13]: 170–172 This, too, is known for many viral infections, and knowledge of the length of both periods is important in the control of outbreaks.[13]: 272 When outbreaks cause an unusually high proportion of cases in a population, community, or region, they are called epidemics. If outbreaks spread worldwide, they are called pandemics.[13]: 891
Epidemics and pandemics
[edit]
A pandemic is a worldwide epidemic. The 1918 flu pandemic, which lasted until 1919, was a category 5 influenza pandemic caused by an unusually severe and deadly influenza A virus. The victims were often healthy young adults, in contrast to most influenza outbreaks, which predominantly affect juvenile, elderly, or otherwise-weakened patients.[26]: 409–415 Older estimates say it killed 40–50 million people,[123] while more recent research suggests that it may have killed as many as 100 million people, or 5% of the world's population in 1918.[124]
Although viral pandemics are rare events, HIV—which evolved from viruses found in monkeys and chimpanzees—has been pandemic since at least the 1980s.[125] During the 20th century there were four pandemics caused by influenza virus and those that occurred in 1918, 1957 and 1968 were severe.[126] Most researchers believe that HIV originated in sub-Saharan Africa during the 20th century;[127] it is now a pandemic, with an estimated 37.9 million people now living with the disease worldwide.[128] There were about 770,000 deaths from AIDS in 2018.[129] The Joint United Nations Programme on HIV/AIDS (UNAIDS) and the World Health Organization (WHO) estimate that AIDS has killed more than 25 million people since it was first recognised on 5 June 1981, making it one of the most destructive epidemics in recorded history.[130] In 2007 there were 2.7 million new HIV infections and 2 million HIV-related deaths.[131]
Several highly lethal viral pathogens are members of the Filoviridae. Filoviruses are filament-like viruses that cause viral hemorrhagic fever, and include ebolaviruses and marburgviruses. Marburg virus, first discovered in 1967, attracted widespread press attention in April 2005 for an outbreak in Angola.[132] Ebola virus disease has also caused intermittent outbreaks with high mortality rates since 1976 when it was first identified. The worst and most recent one is the 2013–2016 West Africa epidemic.[133]
Except for smallpox, most pandemics are caused by newly evolved viruses. These "emergent" viruses are usually mutants of less harmful viruses that have circulated previously either in humans or other animals.[134]
Severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) are caused by new types of coronaviruses. Other coronaviruses are known to cause mild infections in humans,[135] so the virulence and rapid spread of SARS infections—that by July 2003 had caused around 8,000 cases and 800 deaths—was unexpected and most countries were not prepared.[136]
A related coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-Cov-2), thought to have originated in bats, emerged in Wuhan, China in November 2019 and spread rapidly around the world. Infections with the virus caused the COVID-19 pandemic that started in 2020.[94][137][138] Unprecedented restrictions in peacetime were placed on international travel,[139] and curfews were imposed in several major cities worldwide in response to the pandemic.[140]
Cancer
[edit]Viruses are an established cause of cancer in humans and other species. Viral cancers occur only in a minority of infected persons (or animals). Cancer viruses come from a range of virus families, including both RNA and DNA viruses, and so there is no single type of "oncovirus" (an obsolete term originally used for acutely transforming retroviruses). The development of cancer is determined by a variety of factors such as host immunity[141] and mutations in the host.[142] Viruses accepted to cause human cancers include some genotypes of human papillomavirus, hepatitis B virus, hepatitis C virus, Epstein–Barr virus, Kaposi's sarcoma-associated herpesvirus and human T-lymphotropic virus. The most recently discovered human cancer virus is a polyomavirus (Merkel cell polyomavirus) that causes most cases of a rare form of skin cancer called Merkel cell carcinoma.[143] Hepatitis viruses can develop into a chronic viral infection that leads to liver cancer.[144][145] Infection by human T-lymphotropic virus can lead to tropical spastic paraparesis and adult T-cell leukaemia.[146] Human papillomaviruses are an established cause of cancers of cervix, skin, anus, and penis.[147] Within the Herpesviridae, Kaposi's sarcoma-associated herpesvirus causes Kaposi's sarcoma and body-cavity lymphoma, and Epstein–Barr virus causes Burkitt's lymphoma, Hodgkin's lymphoma, B lymphoproliferative disorder, and nasopharyngeal carcinoma.[148] Merkel cell polyomavirus closely related to SV40 and mouse polyomaviruses that have been used as animal models for cancer viruses for over 50 years.[149]
Host defence mechanisms
[edit]The body's first line of defence against viruses is the innate immune system. This comprises cells and other mechanisms that defend the host from infection in a non-specific manner. This means that the cells of the innate system recognise, and respond to, pathogens in a generic way, but, unlike the adaptive immune system, it does not confer long-lasting or protective immunity to the host.[150]
RNA interference is an important innate defence against viruses.[151] Many viruses have a replication strategy that involves double-stranded RNA (dsRNA). When such a virus infects a cell, it releases its RNA molecule or molecules, which immediately bind to a protein complex called a dicer that cuts the RNA into smaller pieces. A biochemical pathway—the RISC complex—is activated, which ensures cell survival by degrading the viral mRNA. Rotaviruses have evolved to avoid this defence mechanism by not uncoating fully inside the cell, and releasing newly produced mRNA through pores in the particle's inner capsid. Their genomic dsRNA remains protected inside the core of the virion.[152][153]
When the adaptive immune system of a vertebrate encounters a virus, it produces specific antibodies that bind to the virus and often render it non-infectious. This is called humoral immunity. Two types of antibodies are important. The first, called IgM, is highly effective at neutralising viruses but is produced by the cells of the immune system only for a few weeks. The second, called IgG, is produced indefinitely. The presence of IgM in the blood of the host is used to test for acute infection, whereas IgG indicates an infection sometime in the past.[154] IgG antibody is measured when tests for immunity are carried out.[155]
Antibodies can continue to be an effective defence mechanism even after viruses have managed to gain entry to the host cell. A protein that is in cells, called TRIM21, can attach to the antibodies on the surface of the virus particle. This primes the subsequent destruction of the virus by the enzymes of the cell's proteosome system.[156]

A second defence of vertebrates against viruses is called cell-mediated immunity and involves immune cells known as T cells. The body's cells constantly display short fragments of their proteins on the cell's surface, and, if a T cell recognises a suspicious viral fragment there, the host cell is destroyed by 'killer T' cells and the virus-specific T-cells proliferate. Cells such as the macrophage are specialists at this antigen presentation.[157] The production of interferon is an important host defence mechanism. This is a hormone produced by the body when viruses are present. Its role in immunity is complex; it eventually stops the viruses from reproducing by killing the infected cell and its close neighbours.[158]
Not all virus infections produce a protective immune response in this way. HIV evades the immune system by constantly changing the amino acid sequence of the proteins on the surface of the virion. This is known as "escape mutation" as the viral epitopes escape recognition by the host immune response. These persistent viruses evade immune control by sequestration, blockade of antigen presentation, cytokine resistance, evasion of natural killer cell activities, escape from apoptosis, and antigenic shift.[159] Other viruses, called 'neurotropic viruses', are disseminated by neural spread where the immune system may be unable to reach them due to immune privilege.[160]
Prevention and treatment
[edit]Because viruses use vital metabolic pathways within host cells to replicate, they are difficult to eliminate without using drugs that cause toxic effects to host cells in general. The most effective medical approaches to viral diseases are vaccinations to provide immunity to infection, and antiviral drugs that selectively interfere with viral replication.
Vaccines
[edit]Vaccination is a cheap and effective way of preventing infections by viruses. Vaccines were used to prevent viral infections long before the discovery of the actual viruses. Their use has resulted in a dramatic decline in morbidity (illness) and mortality (death) associated with viral infections such as polio, measles, mumps and rubella.[161] Smallpox infections have been eradicated.[162] Vaccines are available to prevent over thirteen viral infections of humans,[163] and more are used to prevent viral infections of animals.[164] Vaccines can consist of live-attenuated or killed viruses, viral proteins (antigens), or RNA.[165][166] Live vaccines contain weakened forms of the virus, which do not cause the disease but, nonetheless, confer immunity. Such viruses are called attenuated. Live vaccines can be dangerous when given to people with a weak immunity (who are described as immunocompromised), because in these people, the weakened virus can cause the original disease.[167] Biotechnology and genetic engineering techniques are used to produce subunit vaccines. These vaccines use only the capsid proteins of the virus. Hepatitis B vaccine is an example of this type of vaccine.[168] Subunit vaccines are safe for immunocompromised patients because they cannot cause the disease.[169] The yellow fever virus vaccine, a live-attenuated strain called 17D, is probably the safest and most effective vaccine ever generated.[170]
Antiviral drugs
[edit]
Antiviral drugs are often nucleoside analogues (fake DNA building-blocks), which viruses mistakenly incorporate into their genomes during replication.[171] The life-cycle of the virus is then halted because the newly synthesised DNA is inactive. This is because these analogues lack the hydroxyl groups, which, along with phosphorus atoms, link together to form the strong "backbone" of the DNA molecule. This is called DNA chain termination.[172] Examples of nucleoside analogues are aciclovir for Herpes simplex virus infections and lamivudine for HIV and hepatitis B virus infections. Aciclovir is one of the oldest and most frequently prescribed antiviral drugs.[173] Other antiviral drugs in use target different stages of the viral life cycle. HIV is dependent on a proteolytic enzyme called the HIV-1 protease for it to become fully infectious. There is a large class of drugs called protease inhibitors that inactivate this enzyme.[174] There are around thirteen classes of antiviral drugs each targeting different viruses or stages of viral replication.[171]
Hepatitis C is caused by an RNA virus. In 80% of people infected, the disease is chronic, and without treatment, they are infected for the remainder of their lives. There are effective treatments that use direct-acting antivirals.[175] The treatment of chronic carriers of the hepatitis B virus has also been developed by using similar strategies that include lamivudine and other anti-viral drugs.[176]
Infection in other species
[edit]Viruses infect all cellular life and, although viruses occur universally, each cellular species has its own specific range that often infects only that species.[6]: 3 Some viruses, called satellites, can replicate only within cells that have already been infected by another virus.[30]
Animal viruses
[edit]Viruses are important pathogens of livestock. Diseases such as foot-and-mouth disease and bluetongue are caused by viruses.[177] Companion animals such as cats, dogs, and horses, if not vaccinated, are susceptible to serious viral infections. Canine parvovirus is caused by a small DNA virus and infections are often fatal in pups.[178] Like all invertebrates, the honey bee is susceptible to many viral infections.[179] Most viruses co-exist harmlessly in their host and cause no signs or symptoms of disease.[6]: 4
Plant viruses
[edit]
There are many types of plant viruses, but often they cause only a loss of yield, and it is not economically viable to try to control them. Plant viruses are often spread from plant to plant by organisms, known as vectors. These are usually insects, but some fungi, nematode worms, single-celled organisms, and parasitic plants are vectors.[180] When control of plant virus infections is considered economical, for perennial fruits, for example, efforts are concentrated on killing the vectors and removing alternate hosts such as weeds.[13]: 802 Plant viruses cannot infect humans and other animals because they can reproduce only in living plant cells.[13]: 799–807
Originally from Peru, the potato has become a staple crop worldwide.[181] The potato virus Y causes disease in potatoes and related species including tomatoes and peppers. In the 1980s, this virus acquired economical importance when it proved difficult to control in seed potato crops. Transmitted by aphids, this virus can reduce crop yields by up to 80 per cent, causing significant losses to potato yields.[182]
Plants have elaborate and effective defence mechanisms against viruses. One of the most effective is the presence of so-called resistance (R) genes. Each R gene confers resistance to a particular virus by triggering localised areas of cell death around the infected cell, which can often be seen with the unaided eye as large spots. This stops the infection from spreading.[183] RNA interference is also an effective defence in plants.[13]: 809 When they are infected, plants often produce natural disinfectants that kill viruses, such as salicylic acid, nitric oxide, and reactive oxygen molecules.[184]
Plant virus particles or virus-like particles (VLPs) have applications in both biotechnology and nanotechnology. The capsids of most plant viruses are simple and robust structures and can be produced in large quantities either by the infection of plants or by expression in a variety of heterologous systems. Plant virus particles can be modified genetically and chemically to encapsulate foreign material and can be incorporated into supramolecular structures for use in biotechnology.[185]
Bacterial viruses
[edit]
Bacteriophages are a common and diverse group of viruses and are the most abundant biological entity in aquatic environments—there are up to ten times more of these viruses in the oceans than there are bacteria,[186] reaching levels of 250,000,000 bacteriophages per millilitre of seawater.[187] These viruses infect specific bacteria by binding to surface receptor molecules and then entering the cell. Within a short amount of time, in some cases, just minutes, bacterial polymerase starts translating viral mRNA into protein. These proteins go on to become either new virions within the cell, helper proteins, which help assembly of new virions, or proteins involved in cell lysis. Viral enzymes aid in the breakdown of the cell membrane, and, in the case of the T4 phage, in just over twenty minutes after injection over three hundred phages could be released.[13]: 834–835
The major way bacteria defend themselves from bacteriophages is by producing enzymes that destroy foreign DNA. These enzymes, called restriction endonucleases, cut up the viral DNA that bacteriophages inject into bacterial cells.[188] Bacteria also contain a system that uses CRISPR sequences to retain fragments of the genomes of viruses that the bacteria have come into contact with previously, which allows them to block the virus's replication through a form of RNA interference.[189][190] This genetic system provides bacteria with acquired immunity to infection.[191]
Some bacteriophages are called "temperate" because they cause latent infections and do not immediately destroy their host cells. Instead, their DNA is incorporated with the host cell's as a prophage. These latent infections become productive when the prophage DNA is activated by stimuli such as changes in the environment.[192] The intestines of animals, including humans, contain temperate bacteriophages, which are activated by various stimuli including changes in diet and antibiotics.[193] Although first observed in bacteriophages, many other viruses are known to form proviruses including HIV.[192][194]
Archaeal viruses
[edit]Some viruses replicate within archaea: these are DNA viruses with unusual and sometimes unique shapes.[4][55] These viruses have been studied in most detail in the thermophilic archaea, particularly the orders Sulfolobales and Thermoproteales.[195] Defences against these viruses involve RNA interference from repetitive DNA sequences within archaean genomes that are related to the genes of the viruses.[196][197] Most archaea have CRISPR–Cas systems as an adaptive defence against viruses. These enable archaea to retain sections of viral DNA, which are then used to target and eliminate subsequent infections by the virus using a process similar to RNA interference.[198]
Role in aquatic ecosystems
[edit]Viruses are the most abundant biological entity in aquatic environments.[2] There are about ten million of them in a teaspoon of seawater.[199] Most of these viruses are bacteriophages infecting heterotrophic bacteria and cyanophages infecting cyanobacteria and they are essential to the regulation of saltwater and freshwater ecosystems.[200] Bacteriophages are harmless to plants and animals, and are essential to the regulation of marine and freshwater ecosystems[201] are important mortality agents of phytoplankton, the base of the foodchain in aquatic environments.[202] They infect and destroy bacteria in aquatic microbial communities, and are one of the most important mechanisms of recycling carbon and nutrient cycling in marine environments. The organic molecules released from the dead bacterial cells stimulate fresh bacterial and algal growth, in a process known as the viral shunt.[203] In particular, lysis of bacteria by viruses has been shown to enhance nitrogen cycling and stimulate phytoplankton growth.[204] Viral activity may also affect the biological pump, the process whereby carbon is sequestered in the deep ocean.[205]
Microorganisms constitute more than 90% of the biomass in the sea. It is estimated that viruses kill approximately 20% of this biomass each day and that there are 10 to 15 times as many viruses in the oceans as there are bacteria and archaea.[206] Viruses are also major agents responsible for the destruction of phytoplankton including harmful algal blooms,[207] The number of viruses in the oceans decreases further offshore and deeper into the water, where there are fewer host organisms.[205]
In January 2018, scientists reported that 800 million viruses, mainly of marine origin, are deposited daily from the Earth's atmosphere onto every square meter of the planet's surface, as the result of a global atmospheric stream of viruses, circulating above the weather system but below the altitude of usual airline travel, distributing viruses around the planet.[208][209]
Like any organism, marine mammals are susceptible to viral infections. In 1988 and 2002, thousands of harbour seals were killed in Europe by phocine distemper virus.[210] Many other viruses, including caliciviruses, herpesviruses, adenoviruses and parvoviruses, circulate in marine mammal populations.[205]
In December 2022, scientists reported the first observation of virovory via an experiment on pond water containing chlorovirus, which commonly infects green algae in freshwater environments. When all other microbial food sources were removed from the water, the ciliate Halteria was observed to have increased in number due to the active consumption of chlorovirus as a food source instead of its typical bacterivore diet.[211][212]
Role in evolution
[edit]Viruses are an important natural means of transferring genes between different species, which increases genetic diversity and drives evolution.[9][213][214] It is thought that viruses played a central role in early evolution, before the diversification of the last universal common ancestor into bacteria, archaea and eukaryotes.[215] Viruses are still one of the largest reservoirs of unexplored genetic diversity on Earth.[205]
Applications
[edit]Life sciences and medicine
[edit]
Viruses are important to the study of molecular and cell biology as they provide simple systems that can be used to manipulate and investigate the functions of cells.[26]: 8 The study and use of viruses have provided valuable information about aspects of cell biology.[216] For example, viruses have been useful in the study of genetics and helped our understanding of the basic mechanisms of molecular genetics, such as DNA replication, transcription, RNA processing, translation, protein transport, and immunology.
Geneticists often use viruses as vectors to introduce genes into cells that they are studying. This is useful for making the cell produce a foreign substance, or to study the effect of introducing a new gene into the genome. Similarly, virotherapy uses viruses as vectors to treat various diseases, as they can specifically target cells and DNA. It shows promising use in the treatment of cancer and in gene therapy. Eastern European scientists have used phage therapy as an alternative to antibiotics for some time, and interest in this approach is increasing, because of the high level of antibiotic resistance now found in some pathogenic bacteria.[217] The expression of heterologous proteins by viruses is the basis of several manufacturing processes that are currently being used for the production of various proteins such as vaccine antigens and antibodies. Industrial processes have been recently developed using viral vectors and several pharmaceutical proteins are currently in pre-clinical and clinical trials.[218]
Virotherapy
[edit]Virotherapy involves the use of genetically modified viruses to treat diseases.[219] Viruses have been modified by scientists to reproduce in cancer cells and destroy them but not infect healthy cells. Talimogene laherparepvec (T-VEC), for example, is a modified herpes simplex virus that has had a gene, which is required for viruses to replicate in healthy cells, deleted and replaced with a human gene (GM-CSF) that stimulates immunity. When this virus infects cancer cells, it destroys them and in doing so the presence the GM-CSF gene attracts dendritic cells from the surrounding tissues of the body. The dendritic cells process the dead cancer cells and present components of them to other cells of the immune system.[220] Having completed successful clinical trials, the virus gained approval for the treatment of melanoma in late 2015.[221] Viruses that have been reprogrammed to kill cancer cells are called oncolytic viruses.[222]
Materials science and nanotechnology
[edit]From the viewpoint of a materials scientist, viruses can be regarded as organic nanoparticles.[223] Their surface carries specific tools that enable them to cross the barriers of their host cells. The size and shape of viruses and the number and nature of the functional groups on their surface are precisely defined. As such, viruses are commonly used in materials science as scaffolds for covalently linked surface modifications. A particular quality of viruses is that they can be tailored by directed evolution. The powerful techniques developed by life sciences are becoming the basis of engineering approaches towards nanomaterials, opening a wide range of applications far beyond biology and medicine.[224] Because of their size, shape, and well-defined chemical structures, viruses have been used as templates for organising materials on the nanoscale. Examples include the work at the Naval Research Laboratory in Washington, D.C., using Cowpea mosaic virus (CPMV) particles to amplify signals in DNA microarray based sensors. In this application, the virus particles separate the fluorescent dyes used for signalling to prevent the formation of non-fluorescent dimers that act as quenchers.[225] Another example is the use of CPMV as a nanoscale breadboard for molecular electronics.[226]
Synthetic viruses
[edit]Many viruses can be synthesised de novo ("from scratch"). The first synthetic virus was created in 2002.[227] Although somewhat of a misconception, it is not the actual virus that is synthesised, but rather its DNA genome (in case of a DNA virus), or a cDNA copy of its genome (in case of RNA viruses). For many virus families the naked synthetic DNA or RNA (once enzymatically converted back from the synthetic cDNA) is infectious when introduced into a cell. That is, they contain all the necessary information to produce new viruses. This technology is now being used to investigate novel vaccine strategies.[228] The ability to synthesise viruses has far-reaching consequences, since viruses can no longer be regarded as extinct, as long as the information of their genome sequence is known and permissive cells are available. As of June 2021, the full-length genome sequences of 11,464 different viruses, including smallpox, are publicly available in an online database maintained by the National Institutes of Health.[229]
Weapons
[edit]The ability of viruses to cause devastating epidemics in human societies has led to the concern that viruses could be weaponised for biological warfare. Further concern was raised by the successful recreation of the infamous 1918 influenza virus in a laboratory.[230] The smallpox virus devastated numerous societies throughout history before its eradication. There are only two centres in the world authorised by the WHO to keep stocks of smallpox virus: the State Research Center of Virology and Biotechnology VECTOR in Russia and the Centers for Disease Control and Prevention in the United States.[231] It may be used as a weapon,[231] as the vaccine for smallpox sometimes had severe side-effects, it is no longer used routinely in any country. Thus, much of the modern human population has almost no established resistance to smallpox and would be vulnerable to the virus.[231]
See also
[edit]- Cross-species transmission
- Glossary of virology
- Law of declining virulence – Disproved hypothesis of epidemiologist Theobald Smith
- Non-cellular life
- Gene transfer agent
- Retrozyme
- Smallest organisms
- Theory of virulence – Theory by biologist Paul W. Ewald
- Viral metagenomics
- Viroplasm
- Zoonosis
References
[edit]- ^ Wu KJ (15 April 2020). "There are more viruses than stars in the universe. Why do only some infect us? – More than a quadrillion quadrillion individual viruses exist on Earth, but most are not poised to hop into humans. Can we find the ones that are?". National Geographic Society. Archived from the original on 15 April 2020. Retrieved 18 May 2020.
- ^ a b c Koonin EV, Senkevich TG, Dolja VV (September 2006). "The ancient Virus World and evolution of cells". Biology Direct. 1 (1): 29. doi:10.1186/1745-6150-1-29. PMC 1594570. PMID 16984643.
- ^ Zimmer C (26 February 2021). "The Secret Life of a Coronavirus - An oily, 100-nanometer-wide bubble of genes has killed more than two million people and reshaped the world. Scientists don't quite know what to make of it". The New York Times. Archived from the original on 28 December 2021. Retrieved 28 February 2021.
- ^ a b Lawrence CM, Menon S, Eilers BJ, Bothner B, Khayat R, Douglas T, et al. (May 2009). "Structural and functional studies of archaeal viruses". The Journal of Biological Chemistry. 284 (19): 12599–603. doi:10.1074/jbc.R800078200. PMC 2675988. PMID 19158076.
- ^ Edwards RA, Rohwer F (June 2005). "Viral metagenomics". Nature Reviews. Microbiology. 3 (6): 504–10. doi:10.1038/nrmicro1163. PMID 15886693. S2CID 8059643.
- ^ a b c d e f g h i j k l m n Dimmock NJ, Easton AJ, Leppard K (2007). Introduction to Modern Virology (6th ed.). Blackwell Publishing. ISBN 978-1-4051-3645-7.
- ^ a b c "Virus Taxonomy: 2024 Release". talk.ictvonline.org. International Committee on Taxonomy of Viruses. Retrieved 10 March 2025.
- ^ a b Breitbart M, Rohwer F (June 2005). "Here a virus, there a virus, everywhere the same virus?". Trends in Microbiology. 13 (6): 278–84. doi:10.1016/j.tim.2005.04.003. PMID 15936660.
- ^ a b Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brüssow H (August 2003). "Phage as agents of lateral gene transfer". Current Opinion in Microbiology. 6 (4): 417–24. doi:10.1016/S1369-5274(03)00086-9. PMID 12941415.
- ^ a b Rybicki EP (1990). "The classification of organisms at the edge of life, or problems with virus systematics". South African Journal of Science. 86: 182–86.
- ^ a b Koonin EV, Starokadomskyy P (October 2016). "Are viruses alive? The replicator paradigm sheds decisive light on an old but misguided question". Studies in History and Philosophy of Biological and Biomedical Sciences. 59: 125–34. doi:10.1016/j.shpsc.2016.02.016. PMC 5406846. PMID 26965225.
- ^ Robilotti E, Deresinski S, Pinsky BA (January 2015). "Norovirus". Clinical Microbiology Reviews. 28 (1): 134–64. doi:10.1128/CMR.00075-14. PMC 4284304. PMID 25567225.
- ^ a b c d e f g h i j k l m n o p q r s Shors T (2017). Understanding Viruses. Jones and Bartlett Publishers. ISBN 978-1-284-02592-7.
- ^ "Virus, n.". OED Online. Oxford University Press. March 2015.
- ^ a b Harper D (2011). "virus". The Online Etymology Dictionary. Retrieved 19 December 2014.
- ^ "Virulent, adj.". OED Online. Oxford University Press. March 2015.
- ^ Harper D (2011). "virulent". The Online Etymology Dictionary. Retrieved 19 December 2014.
- ^ Buschard K, Thon R (2003). "Diabetic Animal Models". In Hau J, Van Hoosier Jr GL (eds.). Handbook of Laboratory Animal Science. Animal Models. Vol. II (2nd ed.). CRC Press. pp. 163, 166.
- ^ William T. Stearn: Botanical Latin. History, Grammar, Syntax, Terminology and Vocabulary. David & Charles, 3rd ed., 1983. Quote: "Virus: virus (s.n. II), gen. sing. viri, nom. pl. vira, gen. pl. vīrorum (to be distinguished from virorum, of men)."
- ^ Harper D (2011). "viral". The Online Etymology Dictionary. Retrieved 19 December 2014.
- ^ Harper D (2011). "virion". The Online Etymology Dictionary. Retrieved 19 December 2014.
- ^ Casjens S (2010). Mahy BW, Van Regenmortel MH (eds.). Desk Encyclopedia of General Virology. Boston: Academic Press. p. 167. ISBN 978-0-12-375146-1.
- ^ Iyer LM, Balaji S, Koonin EV, Aravind L (April 2006). "Evolutionary genomics of nucleo-cytoplasmic large DNA viruses". Virus Research. 117 (1): 156–84. doi:10.1016/j.virusres.2006.01.009. PMID 16494962.
- ^ a b Sanjuán R, Nebot MR, Chirico N, Mansky LM, Belshaw R (October 2010). "Viral mutation rates". Journal of Virology. 84 (19): 9733–9748. doi:10.1128/JVI.00694-10. PMC 2937809. PMID 20660197.
- ^ Krupovic M, Dolja VV, Koonin EV (July 2019). "Origin of viruses: primordial replicators recruiting capsids from hosts" (PDF). Nature Reviews. Microbiology. 17 (7): 449–58. doi:10.1038/s41579-019-0205-6. PMID 31142823. S2CID 256744818.
- ^ a b c d e f g h i j k l m n o p q r s t u Collier L, Balows A, Sussman M (1998). Mahy B, Collier LA (eds.). Topley and Wilson's Microbiology and Microbial Infections. Virology. Vol. 1 (9th ed.). ISBN 0-340-66316-2.
- ^ a b c d e f g h Mahy WJ, Regenmortel MH, eds. (2009). Desk Encyclopedia of General Virology. Oxford: Academic Press. ISBN 978-0-12-375146-1.
- ^ McClintock B (June 1950). "The origin and behavior of mutable loci in maize". Proceedings of the National Academy of Sciences of the United States of America. 36 (6): 344–55. Bibcode:1950PNAS...36..344M. doi:10.1073/pnas.36.6.344. PMC 1063197. PMID 15430309.
- ^ Tsagris EM, Martínez de Alba AE, Gozmanova M, Kalantidis K (November 2008). "Viroids". Cellular Microbiology. 10 (11): 2168–79. doi:10.1111/j.1462-5822.2008.01231.x. PMID 18764915. S2CID 221581424.
- ^ a b La Scola B, Desnues C, Pagnier I, Robert C, Barrassi L, Fournous G, et al. (September 2008). "The virophage as a unique parasite of the giant mimivirus". Nature. 455 (7209): 100–04. Bibcode:2008Natur.455..100L. doi:10.1038/nature07218. PMID 18690211. S2CID 4422249.
- ^ Dimmock pp. 15–16
- ^ Zerbini FM, Kitajima EW (September 2022). "From Contagium vivum fluidum to Riboviria: A Tobacco Mosaic Virus-Centric History of Virus Taxonomy". Biomolecules. 12 (10): 1363. doi:10.3390/biom12101363. PMC 9599303. PMID 36291572.
- ^ Holmes EC (October 2007). "Viral evolution in the genomic age". PLOS Biology. 5 (10): e278. doi:10.1371/journal.pbio.0050278. PMC 1994994. PMID 17914905.
- ^ Wimmer E, Mueller S, Tumpey TM, Taubenberger JK (December 2009). "Synthetic viruses: a new opportunity to understand and prevent viral disease". Nature Biotechnology. 27 (12): 1163–72. doi:10.1038/nbt.1593. PMC 2819212. PMID 20010599.
- ^ Horn M (2008). "Chlamydiae as symbionts in eukaryotes". Annual Review of Microbiology. 62: 113–31. doi:10.1146/annurev.micro.62.081307.162818. PMID 18473699.
- ^ Diop A, Raoult D, Fournier PE (February 2019). "Paradoxical evolution of rickettsial genomes". Ticks and Tick-borne Diseases. 10 (2): 462–469. doi:10.1016/j.ttbdis.2018.11.007. PMID 30448253.
- ^ Forterre, P. The virocell concept and environmental microbiology. ISME J 7, 233–236 (2013). https://doi.org/10.1038/ismej.2012.110
- ^ DeLong JP, Al-Sammak MA, Al-Ameeli ZT, Dunigan DD, Edwards KF, Fuhrmann JJ, et al. (February 2022). "Towards an integrative view of virus phenotypes". Nature Reviews. Microbiology. 20 (2): 83–94. doi:10.1038/s41579-021-00612-w. PMID 34522049.
- ^ Krasner R (2014). The microbial challenge: a public health perspective. Burlington, Mass: Jones & Bartlett Learning. ISBN 978-1-4496-7375-8. OCLC 794228026.
- ^ Kiselev NA, Sherman MB, Tsuprun VL (1990). "Negative staining of proteins". Electron Microscopy Reviews. 3 (1): 43–72. doi:10.1016/0892-0354(90)90013-I. PMID 1715774.
- ^ Caspar DL, Klug A (1962). "Physical principles in the construction of regular viruses". Cold Spring Harbor Symposia on Quantitative Biology. 27: 1–24. doi:10.1101/sqb.1962.027.001.005. PMID 14019094.
- ^ Crick FH, Watson JD (March 1956). "Structure of small viruses". Nature. 177 (4506): 473–75. Bibcode:1956Natur.177..473C. doi:10.1038/177473a0. PMID 13309339. S2CID 5740221.
- ^ Falvo MR, Washburn S, Superfine R, Finch M, Brooks FP, Chi V, et al. (March 1997). "Manipulation of individual viruses: friction and mechanical properties". Biophysical Journal. 72 (3): 1396–403. Bibcode:1997BpJ....72.1396F. doi:10.1016/S0006-3495(97)78786-1. PMC 1184522. PMID 9138585.
- ^ Kuznetsov YG, Malkin AJ, Lucas RW, Plomp M, McPherson A (September 2001). "Imaging of viruses by atomic force microscopy". The Journal of General Virology. 82 (Pt 9): 2025–34. doi:10.1099/0022-1317-82-9-2025. PMID 11514711.
- ^ Straus SK, Bo HE (2018). "Filamentous Bacteriophage Proteins and Assembly". Virus Protein and Nucleoprotein Complexes. Subcellular Biochemistry. Vol. 88. pp. 261–79. doi:10.1007/978-981-10-8456-0_12. ISBN 978-981-10-8455-3. PMID 29900501.
- ^ Wilson DP (2016). "Protruding Features of Viral Capsids Are Clustered on Icosahedral Great Circles". PLOS ONE. 11 (4) e0152319. Bibcode:2016PLoSO..1152319W. doi:10.1371/journal.pone.0152319. PMC 4821576. PMID 27045511.
- ^ Casens S (2009). Desk Encyclopedia of General Virology. Boston: Academic Press. pp. 167–74. ISBN 978-0-12-375146-1.
- ^ Dhama K, Khan S, Tiwari R, Sircar S, Bhat S, Malik YS, et al. (September 2020). "Coronavirus Disease 2019-COVID-19". Clinical Microbiology Reviews. 33 (4). doi:10.1128/CMR.00028-20. PMC 7405836. PMID 32580969.
- ^ Rossmann MG, Mesyanzhinov VV, Arisaka F, Leiman PG (April 2004). "The bacteriophage T4 DNA injection machine". Current Opinion in Structural Biology. 14 (2): 171–80. doi:10.1016/j.sbi.2004.02.001. PMID 15093831.
- ^ Long GW, Nobel J, Murphy FA, Herrmann KL, Lourie B (September 1970). "Experience with electron microscopy in the differential diagnosis of smallpox". Applied Microbiology. 20 (3): 497–504. doi:10.1128/AEM.20.3.497-504.1970. PMC 376966. PMID 4322005.
- ^ Suzan-Monti M, La Scola B, Raoult D (April 2006). "Genomic and evolutionary aspects of Mimivirus". Virus Research. 117 (1): 145–55. doi:10.1016/j.virusres.2005.07.011. PMID 16181700.
- ^ Arslan D, Legendre M, Seltzer V, Abergel C, Claverie JM (October 2011). "Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae". Proceedings of the National Academy of Sciences of the United States of America. 108 (42): 17486–91. Bibcode:2011PNAS..10817486A. doi:10.1073/pnas.1110889108. PMC 3198346. PMID 21987820.
- ^ a b Philippe N, Legendre M, Doutre G, Couté Y, Poirot O, Lescot M, et al. (July 2013). "Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes" (PDF). Science. 341 (6143): 281–86. Bibcode:2013Sci...341..281P. doi:10.1126/science.1239181. PMID 23869018. S2CID 16877147.
- ^ Brandes N, Linial M (April 2019). "Giant Viruses-Big Surprises". Viruses. 11 (5): 404. doi:10.3390/v11050404. PMC 6563228. PMID 31052218.
- ^ a b Prangishvili D, Forterre P, Garrett RA (November 2006). "Viruses of the Archaea: a unifying view". Nature Reviews. Microbiology. 4 (11): 837–48. doi:10.1038/nrmicro1527. PMID 17041631. S2CID 9915859.
- ^ "NCBI Viral Genome database". ncbi.nlm.nih.gov. Retrieved 15 January 2017.
- ^ Pennisi E (March 2011). "Microbiology. Going viral: exploring the role of viruses in our bodies". Science. 331 (6024): 1513. Bibcode:2011Sci...331.1513P. doi:10.1126/science.331.6024.1513. PMID 21436418.
- ^ Shi M, Lin XD, Tian JH, Chen LJ, Chen X, Li CX, et al. (December 2016). "Redefining the invertebrate RNA virosphere". Nature. 540 (7634): 539–43. Bibcode:2016Natur.540..539S. doi:10.1038/nature20167. PMID 27880757. S2CID 1198891.
- ^ Levintov L, Vashisth H (January 2024). "Structural and computational studies of HIV-1 RNA". RNA Biology. 21 (1): 167–198. doi:10.1080/15476286.2023.2289709. PMC 10730233. PMID 38100535.
- ^ Saunders VA, Carter J (2007). Virology: principles and applications. Chichester: John Wiley & Sons. p. 72. ISBN 978-0-470-02387-7.
- ^ Belyi VA, Levine AJ, Skalka AM (December 2010). "Sequences from ancestral single-stranded DNA viruses in vertebrate genomes: the parvoviridae and circoviridae are more than 40 to 50 million years old". Journal of Virology. 84 (23): 12458–62. doi:10.1128/JVI.01789-10. PMC 2976387. PMID 20861255.
- ^ Brandes N, Linial M (May 2016). "Gene overlapping and size constraints in the viral world". Biology Direct. 11 (1) 26. doi:10.1186/s13062-016-0128-3. PMC 4875738. PMID 27209091.
- ^ Pressing J, Reanney DC (1984). "Divided genomes and intrinsic noise". Journal of Molecular Evolution. 20 (2): 135–46. Bibcode:1984JMolE..20..135P. doi:10.1007/BF02257374. PMC 7087551. PMID 6433032.
- ^ Duffy S, Holmes EC (June 2009). "Validation of high rates of nucleotide substitution in geminiviruses: phylogenetic evidence from East African cassava mosaic viruses". The Journal of General Virology. 90 (Pt 6): 1539–47. doi:10.1099/vir.0.009266-0. PMC 4091138. PMID 19264617.
- ^ Sandbulte MR, Westgeest KB, Gao J, Xu X, Klimov AI, Russell CA, et al. (December 2011). "Discordant antigenic drift of neuraminidase and hemagglutinin in H1N1 and H3N2 influenza viruses". Proceedings of the National Academy of Sciences of the United States of America. 108 (51): 20748–53. Bibcode:2011PNAS..10820748S. doi:10.1073/pnas.1113801108. PMC 3251064. PMID 22143798.
- ^ Moss RB, Davey RT, Steigbigel RT, Fang F (June 2010). "Targeting pandemic influenza: a primer on influenza antivirals and drug resistance". The Journal of Antimicrobial Chemotherapy. 65 (6): 1086–93. doi:10.1093/jac/dkq100. PMID 20375034.
- ^ Hampson AW, Mackenzie JS (November 2006). "The influenza viruses". The Medical Journal of Australia. 185 (S10): S39–43. doi:10.5694/j.1326-5377.2006.tb00705.x. PMID 17115950. S2CID 17069567.
- ^ Metzner KJ (December 2006). "Detection and significance of minority quasispecies of drug-resistant HIV-1". Journal of HIV Therapy. 11 (4): 74–81. PMID 17578210.
- ^ Goudsmit, Jaap. Viral Sex. Oxford Univ Press, 1998. ISBN 978-0-19-512496-5
- ^ Worobey M, Holmes EC (October 1999). "Evolutionary aspects of recombination in RNA viruses". The Journal of General Virology. 80 (10): 2535–43. doi:10.1099/0022-1317-80-10-2535. PMID 10573145.
- ^ Lukashev AN (2005). "Role of recombination in evolution of enteroviruses". Reviews in Medical Virology. 15 (3): 157–67. doi:10.1002/rmv.457. PMID 15578739. S2CID 26000112.
- ^ Umene K (July 1999). "Mechanism and application of genetic recombination in herpesviruses". Reviews in Medical Virology. 9 (3): 171–82. doi:10.1002/(SICI)1099-1654(199907/09)9:3<171::AID-RMV243>3.0.CO;2-A. PMID 10479778. S2CID 43110533.
- ^ Su S, Wong G, Shi W, Liu J, Lai AC, Zhou J, et al. (June 2016). "Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses". Trends in Microbiology. 24 (6): 490–502. doi:10.1016/j.tim.2016.03.003. PMC 7125511. PMID 27012512.
- ^ Barr JN, Fearns R (June 2010). "How RNA viruses maintain their genome integrity". The Journal of General Virology. 91 (Pt 6): 1373–87. doi:10.1099/vir.0.020818-0. PMID 20335491.
- ^ Freed EO (August 2015). "HIV-1 assembly, release and maturation". Nature Reviews. Microbiology. 13 (8): 484–96. doi:10.1038/nrmicro3490. PMC 6936268. PMID 26119571.
- ^ Yin J, Redovich J (June 2018). "Kinetic Modeling of Virus Growth in Cells". Microbiology and Molecular Biology Reviews. 82 (2). doi:10.1128/MMBR.00066-17. PMC 5968458. PMID 29592895.
- ^ Más V, Melero JA (2013). "Entry of Enveloped Viruses into Host Cells: Membrane Fusion". Structure and Physics of Viruses. Subcellular Biochemistry. Vol. 68. pp. 467–87. doi:10.1007/978-94-007-6552-8_16. ISBN 978-94-007-6551-1. PMC 7121288. PMID 23737062.
- ^ Boevink P, Oparka KJ (August 2005). "Virus-host interactions during movement processes". Plant Physiology. 138 (4): 1815–21. doi:10.1104/pp.105.066761. PMC 1183373. PMID 16172094.
- ^ Blaas D (May 2016). "Viral entry pathways: the example of common cold viruses". Wiener Medizinische Wochenschrift. 166 (7–8): 211–26. doi:10.1007/s10354-016-0461-2. PMC 4871925. PMID 27174165.
- ^ Isomura H, Stinski MF (February 2013). "Coordination of late gene transcription of human cytomegalovirus with viral DNA synthesis: recombinant viruses as potential therapeutic vaccine candidates". Expert Opinion on Therapeutic Targets. 17 (2): 157–66. doi:10.1517/14728222.2013.740460. PMID 23231449. S2CID 11448687.
- ^ Barman S, Ali A, Hui EK, Adhikary L, Nayak DP (September 2001). "Transport of viral proteins to the apical membranes and interaction of matrix protein with glycoproteins in the assembly of influenza viruses". Virus Research. 77 (1): 61–69. doi:10.1016/S0168-1702(01)00266-0. PMID 11451488.
- ^ Staginnus C, Richert-Pöggeler KR (October 2006). "Endogenous pararetroviruses: two-faced travelers in the plant genome". Trends in Plant Science. 11 (10): 485–91. Bibcode:2006TPS....11..485S. doi:10.1016/j.tplants.2006.08.008. PMID 16949329.
- ^ Roulston A, Marcellus RC, Branton PE (1999). "Viruses and apoptosis". Annual Review of Microbiology. 53: 577–628. doi:10.1146/annurev.micro.53.1.577. PMID 10547702.
- ^ Alwine JC (2008). "Modulation of Host Cell Stress Responses by Human Cytomegalovirus". Human Cytomegalovirus. Current Topics in Microbiology and Immunology. Vol. 325. pp. 263–79. doi:10.1007/978-3-540-77349-8_15. ISBN 978-3-540-77348-1. PMID 18637511.
- ^ Barozzi P, Potenza L, Riva G, Vallerini D, Quadrelli C, Bosco R, et al. (December 2007). "B cells and herpesviruses: a model of lymphoproliferation". Autoimmunity Reviews. 7 (2): 132–36. doi:10.1016/j.autrev.2007.02.018. PMID 18035323.
- ^ Subramanya D, Grivas PD (November 2008). "HPV and cervical cancer: updates on an established relationship". Postgraduate Medicine. 120 (4): 7–13. doi:10.3810/pgm.2008.11.1928. PMID 19020360. S2CID 1399003.
- ^ Sinclair J (March 2008). "Human cytomegalovirus: Latency and reactivation in the myeloid lineage". Journal of Clinical Virology. 41 (3): 180–85. doi:10.1016/j.jcv.2007.11.014. PMID 18164651.
- ^ Jordan MC, Jordan GW, Stevens JG, Miller G (June 1984). "Latent herpesviruses of humans". Annals of Internal Medicine. 100 (6): 866–80. doi:10.7326/0003-4819-100-6-866. PMID 6326635.
- ^ Sissons JG, Bain M, Wills MR (February 2002). "Latency and reactivation of human cytomegalovirus". The Journal of Infection. 44 (2): 73–77. doi:10.1053/jinf.2001.0948. PMID 12076064.
- ^ Crawford DH (2011). "Chapter 2: Viruses are Everywhere". Viruses: A Very Short Introduction. Oxford University Press, US. p. 16. ISBN 978-0-19-957485-8.
- ^ Baggesen DL, Sørensen G, Nielsen EM, Wegener HC (January 2010). "Phage typing of Salmonella Typhimurium – is it still a useful tool for surveillance and outbreak investigation?". Euro Surveillance. 15 (4): 19471. PMID 20122382. Retrieved 19 December 2014.
- ^ Parker MT (September 2016). "An Ecological Framework of the Human Virome Provides Classification of Current Knowledge and Identifies Areas of Forthcoming Discovery". The Yale Journal of Biology and Medicine. 89 (3): 339–51. PMC 5045143. PMID 27698618.
- ^ Woolhouse M, Scott F, Hudson Z, Howey R, Chase-Topping M (October 2012). "Human viruses: discovery and emergence". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 367 (1604): 2864–71. doi:10.1098/rstb.2011.0354. PMC 3427559. PMID 22966141.
- ^ a b Ashour HM, Elkhatib WF, Rahman MM, Elshabrawy HA (March 2020). "Insights into the Recent 2019 Novel Coronavirus (SARS-CoV-2) in Light of Past Human Coronavirus Outbreaks". Pathogens. 9 (3): 186. doi:10.3390/pathogens9030186. PMC 7157630. PMID 32143502.
- ^ Lwoff A, Horne RW, Tournier P (June 1962). "[A virus system]". Comptes Rendus Hebdomadaires des Séances de l'Académie des Sciences (in French). 254: 4225–27. PMID 14467544.
- ^ Lwoff A, Horne R, Tournier P (1962). "A system of viruses". Cold Spring Harbor Symposia on Quantitative Biology. 27: 51–55. doi:10.1101/sqb.1962.027.001.008. PMID 13931895.
- ^ Fauquet CM, Fargette D (August 2005). "International Committee on Taxonomy of Viruses and the 3,142 unassigned species". Virology Journal. 2 64. doi:10.1186/1743-422X-2-64. PMC 1208960. PMID 16105179.
- ^ International Committee on Taxonomy of Viruses Executive Committee (May 2020). "The New Scope of Virus Taxonomy: Partitioning the Virosphere Into 15 Hierarchical Ranks". Nat Microbiol. 5 (5): 668–74. doi:10.1038/s41564-020-0709-x. PMC 7186216. PMID 32341570.
- ^ Khan MK, Alam MM (July 2021). "Norovirus Gastroenteritis Outbreaks, Genomic Diversity and Evolution: An Overview". Mymensingh Medical Journal. 30 (3): 863–73. PMID 34226482.
- ^ Eberle J, Gürtler L (2012). "HIV types, groups, subtypes and recombinant forms: errors in replication, selection pressure and quasispecies". Intervirology. 55 (2): 79–83. doi:10.1159/000331993. PMID 22286874. S2CID 5642060.
- ^ International Committee on Taxonomy of Viruses Executive Committee (May 2020). "The new scope of virus taxonomy: partitioning the virosphere into 15 hierarchical ranks". Nature Microbiology. 5 (5): 668–674. doi:10.1038/s41564-020-0709-x. PMC 7186216. PMID 32341570.
- ^ Delwart EL (2007). "Viral metagenomics". Reviews in Medical Virology. 17 (2): 115–131. doi:10.1002/rmv.532. PMC 7169062. PMID 17295196.
- ^ Temin HM, Baltimore D (1972). "RNA-directed DNA synthesis and RNA tumor viruses". Advances in Virus Research. 17: 129–86. doi:10.1016/S0065-3527(08)60749-6. ISBN 978-0120398171. PMID 4348509.
- ^ Baltimore D (1974). "The strategy of RNA viruses". Harvey Lectures. 70 Series. 70: 57–74. PMID 4377923.
- ^ van Regenmortel MH, Mahy BW (January 2004). "Emerging issues in virus taxonomy". Emerging Infectious Diseases. 10 (1): 8–13. doi:10.3201/eid1001.030279. PMC 3322749. PMID 15078590.
- ^ Mayo MA (1999). "Developments in plant virus taxonomy since the publication of the 6th ICTV Report. International Committee on Taxonomy of Viruses". Archives of Virology. 144 (8): 1659–66. doi:10.1007/s007050050620. PMID 10486120. S2CID 33422303.
- ^ de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H (June 2004). "Classification of papillomaviruses". Virology. 324 (1): 17–27. doi:10.1016/j.virol.2004.03.033. PMID 15183049.
- ^ Fisher B, Harvey RP, Champe PC (2007). "Chapter 33 (Disease summaries)". Lippincott's Illustrated Reviews: Microbiology. Lippincott's Illustrated Reviews Series. Hagerstwon, MD: Lippincott Williams & Wilkins. pp. 367–392. ISBN 978-0-7817-8215-9.
- ^ Komaroff AL (December 2006). "Is human herpesvirus-6 a trigger for chronic fatigue syndrome?". Journal of Clinical Virology. 37 (Suppl 1): S39–46. doi:10.1016/S1386-6532(06)70010-5. PMID 17276367.
- ^ Chen CH, Chiu YL, Wei FC, Koong FJ, Liu HC, Shaw CK, et al. (January 1999). "High seroprevalence of Borna virus infection in schizophrenic patients, family members and mental health workers in Taiwan". Molecular Psychiatry. 4 (1): 33–38. doi:10.1038/sj.mp.4000484. PMID 10089006. S2CID 19830976.
- ^ Margolis TP, Elfman FL, Leib D, Pakpour N, Apakupakul K, Imai Y, et al. (October 2007). "Spontaneous reactivation of herpes simplex virus type 1 in latently infected murine sensory ganglia". Journal of Virology. 81 (20): 11069–74. doi:10.1128/JVI.00243-07. PMC 2045564. PMID 17686862.
- ^ Whitley RJ, Roizman B (May 2001). "Herpes simplex virus infections". Lancet. 357 (9267): 1513–18. doi:10.1016/S0140-6736(00)04638-9. PMID 11377626. S2CID 9854903.
- ^ Barton ES, White DW, Cathelyn JS, Brett-McClellan KA, Engle M, Diamond MS, et al. (May 2007). "Herpesvirus latency confers symbiotic protection from bacterial infection". Nature. 447 (7142): 326–29. Bibcode:2007Natur.447..326B. doi:10.1038/nature05762. PMID 17507983. S2CID 4425405.
- ^ Bertoletti A, Gehring A (October 2007). "Immune response and tolerance during chronic hepatitis B virus infection". Hepatology Research. 37 (Suppl 3): S331–38. doi:10.1111/j.1872-034X.2007.00221.x. PMID 17931183. S2CID 13386004.
- ^ Rodrigues C, Deshmukh M, Jacob T, Nukala R, Menon S, Mehta A (2001). "Significance of HBV DNA by PCR over serological markers of HBV in acute and chronic patients". Indian Journal of Medical Microbiology. 19 (3): 141–44. PMID 17664817.
- ^ Nguyen VT, McLaws ML, Dore GJ (December 2007). "Highly endemic hepatitis B infection in rural Vietnam". Journal of Gastroenterology and Hepatology. 22 (12): 2093–100. doi:10.1111/j.1440-1746.2007.05010.x. PMID 17645465. S2CID 29885790.
- ^ Fowler MG, Lampe MA, Jamieson DJ, Kourtis AP, Rogers MF (September 2007). "Reducing the risk of mother-to-child human immunodeficiency virus transmission: past successes, current progress and challenges, and future directions". American Journal of Obstetrics and Gynecology. 197 (3 Suppl): S3–9. doi:10.1016/j.ajog.2007.06.048. PMID 17825648.
- ^ Sauerbrei A, Wutzler P (December 2000). "The congenital varicella syndrome". Journal of Perinatology. 20 (8 Pt 1): 548–54. doi:10.1038/sj.jp.7200457. PMID 11190597. S2CID 7973561.
- ^ a b Antonovics J, Wilson AJ, Forbes MR, Hauffe HC, Kallio ER, Leggett HC, et al. (May 2017). "The evolution of transmission mode". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 372 (1719). doi:10.1098/rstb.2016.0083. PMC 5352810. PMID 28289251.
- ^ Garnett GP (February 2005). "Role of herd immunity in determining the effect of vaccines against sexually transmitted disease". The Journal of Infectious Diseases. 191 (Suppl 1): S97–106. doi:10.1086/425271. PMID 15627236.
- ^ Platonov AE (2006). "[The influence of weather conditions on the epidemiology of vector-borne diseases by the example of West Nile fever in Russia]". Vestnik Rossiiskoi Akademii Meditsinskikh Nauk (in Russian) (2): 25–29. PMID 16544901.
- ^ Jewell CP, Keeling MJ, Roberts GO (December 2009). "Predicting undetected infections during the 2007 foot-and-mouth disease outbreak". Journal of the Royal Society, Interface. 6 (41): 1145–51. doi:10.1098/rsif.2008.0433. PMC 2817150. PMID 19091686.
- ^ Patterson KD, Pyle GF (1991). "The geography and mortality of the 1918 influenza pandemic". Bulletin of the History of Medicine. 65 (1): 4–21. PMID 2021692.
- ^ Johnson NP, Mueller J (2002). "Updating the accounts: global mortality of the 1918–1920 "Spanish" influenza pandemic". Bulletin of the History of Medicine. 76 (1): 105–15. doi:10.1353/bhm.2002.0022. PMID 11875246. S2CID 22974230.
- ^ Eisinger RW, Fauci AS (March 2018). "1". Emerging Infectious Diseases. 24 (3): 413–16. doi:10.3201/eid2403.171797. PMC 5823353. PMID 29460740.
- ^ Qin Y, Zhao MJ, Tan YY, Li XQ, Zheng JD, Peng ZB, et al. (August 2018). "[History of influenza pandemics in China during the past century]". Zhonghua Liu Xing Bing Xue Za Zhi = Zhonghua Liuxingbingxue Zazhi (in Chinese). 39 (8): 1028–31. doi:10.3760/cma.j.issn.0254-6450.2018.08.003 (inactive 1 July 2025). PMID 30180422.
{{cite journal}}: CS1 maint: DOI inactive as of July 2025 (link) - ^ Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, et al. (February 1999). "Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes". Nature. 397 (6718): 436–41. Bibcode:1999Natur.397..436G. doi:10.1038/17130. PMID 9989410. S2CID 4432185.
- ^ "Fact Sheet" (PDF). UNAIDS.org. 2018. Retrieved 12 December 2019.
- ^ "UN AIDS DATA2019". UNAIDS.org. 2019. Retrieved 5 December 2019.
- ^ Mawar N, Saha S, Pandit A, Mahajan U (December 2005). "The third phase of HIV pandemic: social consequences of HIV/AIDS stigma & discrimination & future needs" (PDF). The Indian Journal of Medical Research. 122 (6): 471–84. PMID 16517997. Archived from the original (PDF) on 4 March 2016. Retrieved 19 December 2014.
- ^ "Status of the global HIV epidemic" (PDF). UNAIDS. 2008. Archived from the original (PDF) on 22 November 2015. Retrieved 19 December 2014.
- ^ Towner JS, Khristova ML, Sealy TK, Vincent MJ, Erickson BR, Bawiec DA, et al. (July 2006). "Marburgvirus genomics and association with a large hemorrhagic fever outbreak in Angola". Journal of Virology. 80 (13): 6497–516. doi:10.1128/JVI.00069-06. PMC 1488971. PMID 16775337.
- ^ "World Health Organisation report, 24 September 2014" (PDF).
- ^ "Virology Journal". Virology Journal.
- ^ Weiss SR, Leibowitz JL (2011). Coronavirus pathogenesis. Advances in Virus Research. Vol. 81. pp. 85–164. doi:10.1016/B978-0-12-385885-6.00009-2. ISBN 978-0-12-385885-6. PMC 7149603. PMID 22094080.
- ^ Wong AT, Chen H, Liu SH, Hsu EK, Luk KS, Lai CK, et al. (May 2017). "From SARS to Avian Influenza Preparedness in Hong Kong". Clinical Infectious Diseases. 64 (suppl_2): S98 – S104. doi:10.1093/cid/cix123. PMID 28475794.
- ^ Deng SQ, Peng HJ (February 2020). "Characteristics of and Public Health Responses to the Coronavirus Disease 2019 Outbreak in China". Journal of Clinical Medicine. 9 (2): 575. doi:10.3390/jcm9020575. PMC 7074453. PMID 32093211.
- ^ Han Q, Lin Q, Jin S, You L (April 2020). "Coronavirus 2019-nCoV: A brief perspective from the front line". The Journal of Infection. 80 (4): 373–77. doi:10.1016/j.jinf.2020.02.010. PMC 7102581. PMID 32109444.
- ^ Londoño E, Ortiz A (16 March 2020). "Coronavirus Travel Restrictions, Across the Globe". The New York Times.
- ^ "US takes more big pandemic response steps; Europe COVID-19 cases soar". CIDRAP. 15 March 2020.
- ^ Einstein MH, Schiller JT, Viscidi RP, Strickler HD, Coursaget P, Tan T, et al. (June 2009). "Clinician's guide to human papillomavirus immunology: knowns and unknowns". The Lancet. Infectious Diseases. 9 (6): 347–56. doi:10.1016/S1473-3099(09)70108-2. PMID 19467474.
- ^ Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, et al. (October 2008). "T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus". Proceedings of the National Academy of Sciences of the United States of America. 105 (42): 16272–77. Bibcode:2008PNAS..10516272S. doi:10.1073/pnas.0806526105. PMC 2551627. PMID 18812503.
- ^ Pulitzer MP, Amin BD, Busam KJ (May 2009). "Merkel cell carcinoma: review". Advances in Anatomic Pathology. 16 (3): 135–44. doi:10.1097/PAP.0b013e3181a12f5a. PMID 19395876. S2CID 36110778.
- ^ Koike K (June 2007). "Hepatitis C virus contributes to hepatocarcinogenesis by modulating metabolic and intracellular signaling pathways". Journal of Gastroenterology and Hepatology. 22 (Suppl 1): S108–11. doi:10.1111/j.1440-1746.2006.04669.x. PMID 17567457. S2CID 25399220.
- ^ Hu J, Ludgate L (2007). "HIV–HBV and HIV–HCV Coinfection and Liver Cancer Development". Aids-Associated Viral Oncogenesis. Cancer Treatment and Research. Vol. 133. pp. 241–52. doi:10.1007/978-0-387-46816-7_9. ISBN 978-0-387-46804-4. PMID 17672044.
- ^ Bellon M, Nicot C (2007). "Telomerase: a crucial player in HTLV-I-induced human T-cell leukemia". Cancer Genomics & Proteomics. 4 (1): 21–25. PMID 17726237.
- ^ Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S (September 2007). "Human papillomavirus and cervical cancer". Lancet. 370 (9590): 890–907. doi:10.1016/S0140-6736(07)61416-0. PMID 17826171. S2CID 20196938.
- ^ Klein E, Kis LL, Klein G (February 2007). "Epstein-Barr virus infection in humans: from harmless to life endangering virus-lymphocyte interactions". Oncogene. 26 (9): 1297–305. doi:10.1038/sj.onc.1210240. PMID 17322915.
- ^ Zur Hausen H (July 2008). "Novel human polyomaviruses – re-emergence of a well known virus family as possible human carcinogens". International Journal of Cancer. 123 (2): 247–50. doi:10.1002/ijc.23620. PMID 18449881. S2CID 9482506.
- ^ Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walters P (2002). Molecular Biology of the Cell (4th ed.). New York and London: Garland Science. ISBN 0-8153-3218-1.
- ^ Ding SW, Voinnet O (August 2007). "Antiviral immunity directed by small RNAs". Cell. 130 (3): 413–26. doi:10.1016/j.cell.2007.07.039. PMC 2703654. PMID 17693253.
- ^ Patton JT, Vasquez-Del Carpio R, Spencer E (2004). "Replication and transcription of the rotavirus genome". Current Pharmaceutical Design. 10 (30): 3769–77. doi:10.2174/1381612043382620. PMID 15579070.
- ^ Jayaram H, Estes MK, Prasad BV (April 2004). "Emerging themes in rotavirus cell entry, genome organization, transcription and replication". Virus Research. 101 (1): 67–81. doi:10.1016/j.virusres.2003.12.007. PMID 15010218.
- ^ Greer S, Alexander GJ (December 1995). "Viral serology and detection". Baillière's Clinical Gastroenterology. 9 (4): 689–721. doi:10.1016/0950-3528(95)90057-8. PMID 8903801.
- ^ Matter L, Kogelschatz K, Germann D (April 1997). "Serum levels of rubella virus antibodies indicating immunity: response to vaccination of subjects with low or undetectable antibody concentrations". The Journal of Infectious Diseases. 175 (4): 749–55. doi:10.1086/513967. PMID 9086126.
- ^ Mallery DL, McEwan WA, Bidgood SR, Towers GJ, Johnson CM, James LC (November 2010). "Antibodies mediate intracellular immunity through tripartite motif-containing 21 (TRIM21)". Proceedings of the National Academy of Sciences of the United States of America. 107 (46): 19985–90. Bibcode:2010PNAS..10719985M. doi:10.1073/pnas.1014074107. PMC 2993423. PMID 21045130.
- ^ Cascalho M, Platt JL (2007). "Novel functions of B cells". Critical Reviews in Immunology. 27 (2): 141–51. doi:10.1615/critrevimmunol.v27.i2.20. PMID 17725500.
- ^ Le Page C, Génin P, Baines MG, Hiscott J (2000). "Interferon activation and innate immunity". Reviews in Immunogenetics. 2 (3): 374–86. PMID 11256746.
- ^ Hilleman MR (October 2004). "Strategies and mechanisms for host and pathogen survival in acute and persistent viral infections". Proceedings of the National Academy of Sciences of the United States of America. 101 (Suppl 2): 14560–66. Bibcode:2004PNAS..10114560H. doi:10.1073/pnas.0404758101. PMC 521982. PMID 15297608.
- ^ Libbey JE, Fujinami RS (2014). "Adaptive immune response to viral infections in the central nervous system". Neurovirology. Handbook of Clinical Neurology. Vol. 123. pp. 225–47. doi:10.1016/B978-0-444-53488-0.00010-9. ISBN 978-0444534880. PMC 4370180. PMID 25015488.
- ^ Asaria P, MacMahon E (October 2006). "Measles in the United Kingdom: can we eradicate it by 2010?". BMJ. 333 (7574): 890–95. doi:10.1136/bmj.38989.445845.7C. PMC 1626346. PMID 17068034.
- ^ Lane JM (2006). "Mass vaccination and surveillance/containment in the eradication of smallpox". Mass Vaccination: Global Aspects — Progress and Obstacles. Current Topics in Microbiology and Immunology. Vol. 304. pp. 17–29. doi:10.1007/3-540-36583-4_2. ISBN 978-3-540-29382-8. PMC 7120753. PMID 16989262.
- ^ Arvin AM, Greenberg HB (January 2006). "New viral vaccines". Virology. 344 (1): 240–49. doi:10.1016/j.virol.2005.09.057. PMID 16364754.
- ^ Pastoret PP, Schudel AA, Lombard M (August 2007). "Conclusions--future trends in veterinary vaccinology". Revue Scientifique et Technique. 26 (2): 489–94, 495–501, 503–09. doi:10.20506/rst.26.2.1759. PMID 17892169.
- ^ Palese P (January 2006). "Making better influenza virus vaccines?". Emerging Infectious Diseases. 12 (1): 61–65. doi:10.3201/eid1201.051043. PMC 3291403. PMID 16494719.
- ^ Anand P, Stahel VP (May 2021). "Review the safety of Covid-19 mRNA vaccines: a review". Patient Safety in Surgery. 15 (1): 20. doi:10.1186/s13037-021-00291-9. PMC 8087878. PMID 33933145.
- ^ Thomssen R (1975). "Live attenuated versus killed virus vaccines". Monographs in Allergy. 9: 155–76. PMID 1090805.
- ^ McLean AA (1986). "Development of vaccines against hepatitis A and hepatitis B". Reviews of Infectious Diseases. 8 (4): 591–98. doi:10.1093/clinids/8.4.591. PMID 3018891.
- ^ Casswall TH, Fischler B (October 2005). "Vaccination of the immunocompromised child". Expert Review of Vaccines. 4 (5): 725–38. doi:10.1586/14760584.4.5.725. PMID 16221073. S2CID 40821818.
- ^ Barnett ED, Wilder-Smith A, Wilson ME (July 2008). "Yellow fever vaccines and international travelers". Expert Review of Vaccines. 7 (5): 579–87. doi:10.1586/14760584.7.5.579. PMID 18564013. S2CID 19352868.
- ^ a b De Clercq E, Li G (July 2016). "Approved Antiviral Drugs over the Past 50 Years". Clinical Microbiology Reviews. 29 (3): 695–747. doi:10.1128/CMR.00102-15. PMC 4978613. PMID 27281742.
- ^ Magden J, Kääriäinen L, Ahola T (March 2005). "Inhibitors of virus replication: recent developments and prospects". Applied Microbiology and Biotechnology. 66 (6): 612–21. doi:10.1007/s00253-004-1783-3. PMC 7082807. PMID 15592828.
- ^ Mindel A, Sutherland S (September 1983). "Genital herpes – the disease and its treatment including intravenous acyclovir". The Journal of Antimicrobial Chemotherapy. 12 (Suppl B): 51–59. doi:10.1093/jac/12.suppl_b.51. PMID 6355051.
- ^ Palmisano L, Vella S (2011). "A brief history of antiretroviral therapy of HIV infection: success and challenges". Annali dell'Istituto Superiore di Sanità. 47 (1): 44–48. doi:10.4415/ANN_11_01_10. PMID 21430338.
- ^ Falade-Nwulia O, Suarez-Cuervo C, Nelson DR, Fried MW, Segal JB, Sulkowski MS (May 2017). "Oral Direct-Acting Agent Therapy for Hepatitis C Virus Infection: A Systematic Review". Annals of Internal Medicine. 166 (9): 637–48. doi:10.7326/M16-2575. PMC 5486987. PMID 28319996.
- ^ Nguyen MH, Wong G, Gane E, Kao JH, Dusheiko G (March 2020). "Hepatitis B Virus: Advances in Prevention, Diagnosis, and Therapy". Clinical Microbiology Reviews. 33 (2). doi:10.1128/CMR.00046-19. PMC 7048015. PMID 32102898.
- ^ Goris N, Vandenbussche F, De Clercq K (April 2008). "Potential of antiviral therapy and prophylaxis for controlling RNA viral infections of livestock". Antiviral Research. 78 (1): 170–78. doi:10.1016/j.antiviral.2007.10.003. PMID 18035428.
- ^ Carmichael LE (2005). "An annotated historical account of canine parvovirus". Journal of Veterinary Medicine. B, Infectious Diseases and Veterinary Public Health. 52 (7–8): 303–11. doi:10.1111/j.1439-0450.2005.00868.x. PMID 16316389.
- ^ Chen Y, Zhao Y, Hammond J, Hsu HT, Evans J, Feldlaufer M (October–November 2004). "Multiple virus infections in the honey bee and genome divergence of honey bee viruses". Journal of Invertebrate Pathology. 87 (2–3): 84–93. Bibcode:2004JInvP..87...84C. doi:10.1016/j.jip.2004.07.005. PMID 15579317.
- ^ Hull R (2002). "Chapter 12: Transmission 2: Mechanical, Seed, Pollen and Epidemiology". Matthews' Plant Virology (4th ed.). Academic Press. pp. 555–56. doi:10.1016/B978-012361160-4/50063-3. ISBN 978-0123611604. Retrieved 21 February 2022.
- ^ Zaheer K, Akhtar MH (2016). "Potato Production, Usage, and Nutrition – A Review". Critical Reviews in Food Science and Nutrition. 56 (5): 711–21. doi:10.1080/10408398.2012.724479. PMID 24925679. S2CID 33074838.
- ^ Fuentes S, Jones RA, Matsuoka H, Ohshima K, Kreuze J, Gibbs AJ (July 2019). "Potato virus Y; the Andean connection". Virus Evolution. 5 (2) vez037. doi:10.1093/ve/vez037. PMC 6755682. PMID 31559020.
- ^ Dinesh-Kumar SP, Tham WH, Baker BJ (December 2000). "Structure-function analysis of the tobacco mosaic virus resistance gene N". Proceedings of the National Academy of Sciences of the United States of America. 97 (26): 14789–94. Bibcode:2000PNAS...9714789D. doi:10.1073/pnas.97.26.14789. PMC 18997. PMID 11121079.
- ^ Soosaar JL, Burch-Smith TM, Dinesh-Kumar SP (October 2005). "Mechanisms of plant resistance to viruses". Nature Reviews. Microbiology. 3 (10): 789–98. doi:10.1038/nrmicro1239. PMID 16132037. S2CID 27311732.
- ^ Lomonossoff GP (2011). "Virus Particles and the Uses of Such Particles in Bio- and Nanotechnology". Recent Advances in Plant Virology. Caister Academic Press. ISBN 978-1-904455-75-2.
- ^ Wommack KE, Colwell RR (March 2000). "Virioplankton: viruses in aquatic ecosystems". Microbiology and Molecular Biology Reviews. 64 (1): 69–114. doi:10.1128/MMBR.64.1.69-114.2000. PMC 98987. PMID 10704475.
- ^ Bergh O, Børsheim KY, Bratbak G, Heldal M (August 1989). "High abundance of viruses found in aquatic environments". Nature. 340 (6233): 467–68. Bibcode:1989Natur.340..467B. doi:10.1038/340467a0. PMID 2755508. S2CID 4271861.
- ^ Bickle TA, Krüger DH (June 1993). "Biology of DNA restriction". Microbiological Reviews. 57 (2): 434–50. doi:10.1128/MMBR.57.2.434-450.1993. PMC 372918. PMID 8336674.
- ^ Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. (March 2007). "CRISPR provides acquired resistance against viruses in prokaryotes". Science. 315 (5819): 1709–12. Bibcode:2007Sci...315.1709B. doi:10.1126/science.1138140. hdl:20.500.11794/38902. PMID 17379808. S2CID 3888761.
- ^ Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, et al. (August 2008). "Small CRISPR RNAs guide antiviral defense in prokaryotes". Science. 321 (5891): 960–64. Bibcode:2008Sci...321..960B. doi:10.1126/science.1159689. PMC 5898235. PMID 18703739.
- ^ Mojica FJ, Rodriguez-Valera F (September 2016). "The discovery of CRISPR in archaea and bacteria". The FEBS Journal. 283 (17): 3162–69. doi:10.1111/febs.13766. hdl:10045/57676. PMID 27234458. S2CID 42827598.
- ^ a b Weiss RA (2017). "Exchange of Genetic Sequences Between Viruses and Hosts". Viruses, Genes, and Cancer. Current Topics in Microbiology and Immunology. Vol. 407. pp. 1–29. doi:10.1007/82_2017_21. ISBN 978-3-319-61803-6. PMID 28550453.
- ^ Hu J, Ye H, Wang S, Wang J, Han D (2021). "Prophage Activation in the Intestine: Insights Into Functions and Possible Applications". Frontiers in Microbiology. 12 785634. doi:10.3389/fmicb.2021.785634. PMC 8710666. PMID 34966370.
- ^ Russ E, Iordanskiy S (January 2023). "Endogenous Retroviruses as Modulators of Innate Immunity". Pathogens. 12 (2): 162. doi:10.3390/pathogens12020162. PMC 9963469. PMID 36839434.
- ^ Prangishvili D, Garrett RA (April 2004). "Exceptionally diverse morphotypes and genomes of crenarchaeal hyperthermophilic viruses" (PDF). Biochemical Society Transactions. 32 (Pt 2): 204–08. doi:10.1042/BST0320204. PMID 15046572. S2CID 20018642.
- ^ Mojica FJ, Díez-Villaseñor C, García-Martínez J, Soria E (February 2005). "Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements". Journal of Molecular Evolution. 60 (2): 174–82. Bibcode:2005JMolE..60..174M. doi:10.1007/s00239-004-0046-3. PMID 15791728. S2CID 27481111.
- ^ Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV (March 2006). "A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action". Biology Direct. 1 7. doi:10.1186/1745-6150-1-7. PMC 1462988. PMID 16545108.
- ^ van der Oost J, Westra ER, Jackson RN, Wiedenheft B (July 2014). "Unravelling the structural and mechanistic basis of CRISPR-Cas systems". Nature Reviews. Microbiology. 12 (7): 479–92. doi:10.1038/nrmicro3279. PMC 4225775. PMID 24909109.
- ^ Dávila-Ramos S, Castelán-Sánchez HG, Martínez-Ávila L, Sánchez-Carbente MD, Peralta R, Hernández-Mendoza A, et al. (2019). "A Review on Viral Metagenomics in Extreme Environments". Frontiers in Microbiology. 10: 2403. doi:10.3389/fmicb.2019.02403. PMC 6842933. PMID 31749771.
- ^ Zhang QY, Gui JF (December 2018). "Diversity, evolutionary contribution and ecological roles of aquatic viruses". Science China Life Sciences. 61 (12): 1486–1502. doi:10.1007/s11427-018-9414-7. PMID 30443861. S2CID 53564176.
- ^ Weitz JS, Wilhelm SW (2013). "An ocean of viruses". The Scientist. 27 (7): 35–39.
- ^ Suttle CA (September 2005). "Viruses in the sea". Nature. 437 (7057): 356–61. Bibcode:2005Natur.437..356S. doi:10.1038/nature04160. PMID 16163346. S2CID 4370363.
- ^ Wilhelm SW, Suttle CA (1999). "Viruses and nutrient cycles in the sea: viruses play critical roles in the structure and function of aquatic food webs". BioScience. 49 (10): 781–88. doi:10.2307/1313569. JSTOR 1313569.
- ^ Shelford EJ, Suttle CA (2018). "Virus-mediated transfer of nitrogen from heterotrophic bacteria to phytoplankton". Biogeosciences. 15 (3): 809–15. Bibcode:2018BGeo...15..809S. doi:10.5194/bg-15-809-2018.
- ^ a b c d Suttle CA (October 2007). "Marine viruses – major players in the global ecosystem". Nature Reviews. Microbiology. 5 (10): 801–12. doi:10.1038/nrmicro1750. PMID 17853907. S2CID 4658457.
- ^ Wigington CH, Sonderegger D, Brussaard CP, Buchan A, Finke JF, Fuhrman JA, et al. (January 2016). "Re-examination of the relationship between marine virus and microbial cell abundances" (PDF). Nature Microbiology. 1 (15024) 15024. doi:10.1038/nmicrobiol.2015.24. PMID 27572161. S2CID 52829633.
- ^ Brussaard CP (2004). "Viral control of phytoplankton populations – a review". The Journal of Eukaryotic Microbiology. 51 (2): 125–38. doi:10.1111/j.1550-7408.2004.tb00537.x. PMID 15134247. S2CID 21017882.
- ^ Robbins J (13 April 2018). "Trillions Upon Trillions of Viruses Fall From the Sky Each Day". The New York Times. Retrieved 14 April 2018.
- ^ Reche I, D'Orta G, Mladenov N, Winget DM, Suttle CA (April 2018). "Deposition rates of viruses and bacteria above the atmospheric boundary layer". The ISME Journal. 12 (4): 1154–62. Bibcode:2018ISMEJ..12.1154R. doi:10.1038/s41396-017-0042-4. PMC 5864199. PMID 29379178.
- ^ Hall AJ, Jepson PD, Goodman SJ, Harkonen T (2006). "Phocine distemper virus in the North and European Seas – data and models, nature and nurture". Biological Conservation. 131 (2): 221–29. Bibcode:2006BCons.131..221H. doi:10.1016/j.biocon.2006.04.008.
- ^ DeLong JP, Van Etten JL, Al-Ameeli Z, Agarkova IV, Dunigan DD (January 2023). "The consumption of viruses returns energy to food chains". Proceedings of the National Academy of Sciences of the United States of America. 120 (1) e2215000120. Bibcode:2023PNAS..12015000D. doi:10.1073/pnas.2215000120. PMC 9910503. PMID 36574690. S2CID 255219850.
- ^ Irving M (28 December 2022). "First "virovore" discovered: An organism that eats viruses". New Atlas. Archived from the original on 29 December 2022. Retrieved 29 December 2022.
- ^ Broecker F, Moelling K (2019). "What viruses tell us about evolution and immunity: beyond Darwin?". Annals of the New York Academy of Sciences. 1447 (1): 53–68. Bibcode:2019NYASA1447...53B. doi:10.1111/nyas.14097. PMC 6850104. PMID 31032941.
- ^ Lauer, A. (2021). Viruses, Underestimated Drivers of Ecology and Evolution of Life. In: Hurst, C.J. (eds) Microbes: The Foundation Stone of the Biosphere. Advances in Environmental Microbiology, vol 8. Springer, Cham. https://doi.org/10.1007/978-3-030-63512-1_28
- ^ Forterre P, Philippe H (June 1999). "The last universal common ancestor (LUCA), simple or complex?". The Biological Bulletin. 196 (3): 373–75, discussion 375–77. doi:10.2307/1542973. JSTOR 1542973. PMID 11536914.
- ^ Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J (2000). "Viruses: Structure, Function, and Uses". Molecular Cell Biology (4th ed.). New York: W. H. Freeman.
- ^ Matsuzaki S, Rashel M, Uchiyama J, Sakurai S, Ujihara T, Kuroda M, et al. (October 2005). "Bacteriophage therapy: a revitalized therapy against bacterial infectious diseases". Journal of Infection and Chemotherapy. 11 (5): 211–19. doi:10.1007/s10156-005-0408-9. PMID 16258815. S2CID 8107934.
- ^ Gleba YY, Giritch A (2011). "Plant Viral Vectors for Protein Expression". Recent Advances in Plant Virology. Caister Academic Press. ISBN 978-1-904455-75-2.
- ^ Jefferson A, Cadet VE, Hielscher A (September 2015). "The mechanisms of genetically modified vaccinia viruses for the treatment of cancer". Critical Reviews in Oncology/Hematology. 95 (3): 407–16. doi:10.1016/j.critrevonc.2015.04.001. PMID 25900073.
- ^ Karimkhani C, Gonzalez R, Dellavalle RP (August 2014). "A review of novel therapies for melanoma". American Journal of Clinical Dermatology. 15 (4): 323–37. doi:10.1007/s40257-014-0083-7. PMID 24928310. S2CID 38864550.
- ^ "FDA approves Amgen's injected immunotherapy for melanoma". Reuters. 27 October 2015. Retrieved 24 January 2020.
- ^ Burke J, Nieva J, Borad MJ, Breitbach CJ (August 2015). "Oncolytic viruses: perspectives on clinical development". Current Opinion in Virology. 13: 55–60. doi:10.1016/j.coviro.2015.03.020. PMID 25989094.
- ^ Dogic Z (2016). "Filamentous Phages As a Model System in Soft Matter Physics". Frontiers in Microbiology. 7: 1013. doi:10.3389/fmicb.2016.01013. PMC 4927585. PMID 27446051.
- ^ Fischlechner M, Donath E (2007). "Viruses as building blocks for materials and devices". Angewandte Chemie. 46 (18): 3184–93. doi:10.1002/anie.200603445. PMID 17348058.
- ^ Soto CM, Blum AS, Vora GJ, Lebedev N, Meador CE, Won AP, et al. (April 2006). "Fluorescent signal amplification of carbocyanine dyes using engineered viral nanoparticles". Journal of the American Chemical Society. 128 (15): 5184–89. Bibcode:2006JAChS.128.5184S. doi:10.1021/ja058574x. PMID 16608355.
- ^ Blum AS, Soto CM, Wilson CD, Brower TL, Pollack SK, Schull TL, et al. (July 2005). "An engineered virus as a scaffold for three-dimensional self-assembly on the nanoscale". Small. 1 (7): 702–06. doi:10.1002/smll.200500021. PMID 17193509.
- ^ Cello J, Paul AV, Wimmer E (August 2002). "Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template". Science. 297 (5583): 1016–18. Bibcode:2002Sci...297.1016C. doi:10.1126/science.1072266. PMID 12114528. S2CID 5810309.
- ^ Coleman JR, Papamichail D, Skiena S, Futcher B, Wimmer E, Mueller S (June 2008). "Virus attenuation by genome-scale changes in codon pair bias". Science. 320 (5884): 1784–87. Bibcode:2008Sci...320.1784C. doi:10.1126/science.1155761. PMC 2754401. PMID 18583614.
- ^ "NIH viral genome database". Ncbi.nlm.nih.gov. Retrieved 28 June 2021.
- ^ Zilinskas RA (August 2017). "A brief history of biological weapons programmes and the use of animal pathogens as biological warfare agents". Revue Scientifique et Technique (International Office of Epizootics). 36 (2): 415–22. doi:10.20506/rst.36.2.2662. PMID 30152475.
- ^ a b c Artenstein AW, Grabenstein JD (October 2008). "Smallpox vaccines for biodefense: need and feasibility". Expert Review of Vaccines. 7 (8): 1225–37. doi:10.1586/14760584.7.8.1225. PMC 9709930. PMID 18844596. S2CID 33855724.
External links
[edit]Virus
View on GrokipediaHistory
Etymology
The word virus derives from Latin vīrus, signifying "poison," "sap of plants," or "slimy liquid."[11] This term traces to Proto-Italic *weis-o-(s-), denoting poison, with possible cognates in Old Church Slavonic višnja for cherry, potentially alluding to toxic fruits.[11] In English, virus first appears in the late 14th century, borrowed directly from Latin to describe a poisonous substance, as evidenced in translations like John Trevisa's Bartholomaeus Anglicus (c. 1398). By the 1590s, it extended to "venomous exudation from a living body," and by 1798, metaphorically to an agent causing infectious disease.[11] The modern scientific usage for submicroscopic pathogens causing infection dates to 1883, coinciding with microbiological advances identifying filterable agents beyond bacteria.[11][12] This shift retained the connotation of a noxious, invisible toxin, aligning with empirical observations of contagion without visible organisms.[13]Early Observations
In 1892, Russian microbiologist Dmitri Ivanovsky conducted filtration experiments on tobacco plants affected by mosaic disease, a condition first described by Adolf Mayer in 1879. Using Chamberland porcelain filters with pores small enough to retain bacteria, Ivanovsky found that the filtered sap from diseased leaves remained infectious when applied to healthy plants, indicating the presence of an ultrafilterable agent smaller than known pathogenic bacteria.[14] [15] Ivanovsky initially interpreted this as evidence of a bacterial toxin or dissolved enzymatic substance rather than a discrete living pathogen, as the agent did not form visible colonies in culture media.[16] These findings challenged the prevailing germ theory, which attributed infectious diseases primarily to cultivable bacteria as demonstrated by Robert Koch's postulates in the 1880s. In 1898, Dutch microbiologist Martinus Beijerinck replicated and extended Ivanovsky's work, confirming the filterable nature of the tobacco mosaic agent through serial dilutions and passage experiments that showed progressive dilution reduced infectivity predictably, inconsistent with toxin persistence.[17] Beijerinck proposed the term contagium vivum fluidum ("contagious living fluid") to describe a self-replicating, non-cellular entity that multiplied only within living host cells and could not be grown independently, distinguishing it from bacteria and laying foundational concepts for virology.[17] [18] Concurrently in 1898, German scientists Friedrich Loeffler and Paul Frosch reported similar filtration results for foot-and-mouth disease in cattle, the first such observation in animals. Their experiments demonstrated that the infectious agent passed through bacteria-retaining filters and required living host tissue for propagation, extending the filterable agent paradigm beyond plants.[19] [18] These early observations collectively established that certain diseases were caused by submicroscopic, filter-passing pathogens—later termed viruses—incapable of independent metabolism or growth, prompting a reevaluation of infectious disease causation.[17]Scientific Discovery
In 1892, Russian biologist Dmitri Ivanovsky investigated tobacco mosaic disease by filtering sap from infected plants through porcelain Chamberland filters designed to retain bacteria; the filtrate remained infectious upon inoculation to healthy plants, indicating the causal agent was smaller than known bacteria or possibly a toxin.[14] Ivanovsky initially favored a toxin hypothesis but noted the agent's persistence and infectivity, marking the first experimental evidence of a filterable pathogen.[20] In 1898, Dutch microbiologist Martinus Beijerinck replicated Ivanovsky's filtration experiments and demonstrated that the infectious agent multiplied exponentially in host tissues, diluting and re-concentrating in a manner inconsistent with a static toxin; he termed it contagium vivum fluidum, a self-replicating, fluid infectious principle that required living host cells for propagation, distinguishing it from bacteria.[21] That same year, German scientists Friedrich Loeffler and Paul Frosch applied filtration to foot-and-mouth disease in cattle, identifying the first animal virus and showing it could be transmitted via filtered lymph, extending the concept beyond plants.[22] Advances in the 1930s shifted viruses from abstract entities to tangible macromolecules. In 1935, American biochemist Wendell Stanley isolated and crystallized tobacco mosaic virus (TMV) as a nucleoprotein, demonstrating that purified crystals retained infectivity after redissolution, challenging views of viruses as merely fluids and highlighting their molecular nature akin to enzymes yet capable of self-assembly.[23] [20] Direct visualization emerged with electron microscopy in the late 1930s; in 1939, Helmut Ruska and colleagues imaged TMV particles as rod-shaped structures approximately 300 nm long, confirming their particulate form and submicroscopic size, while 1940 micrographs of bacteriophages revealed tadpole-like morphologies attached to bacteria.[24] These observations solidified viruses as discrete, obligate intracellular parasites, paving the way for virology as a distinct field.[25]Hypotheses on Viral Origins
The origins of viruses remain unresolved, with three principal hypotheses proposed based on comparative genomics, evolutionary modeling, and inferences from viral replication strategies. These include the progressive (or escape) hypothesis, the regressive (or reduction) hypothesis, and the virus-first (or co-evolution) hypothesis. Empirical evidence is indirect, derived primarily from viral genome sequences showing mosaics of cellular-like and unique genes, alongside the absence of viral fossils predating cellular life around 3.5–4 billion years ago. No single hypothesis accounts for the diversity of viral forms, including RNA viruses with high mutation rates and large DNA viruses encoding hundreds of proteins.[9][26] The progressive hypothesis posits that viruses evolved from intragenomic elements, such as plasmids, transposons, or gene fragments, that acquired capsid proteins and egress mechanisms, enabling autonomous transmission between cells. This scenario is supported by the similarity of some viral genes to host mobile elements and the observed integration of viral sequences into eukaryotic genomes, suggesting repeated "escapes" over evolutionary time. For instance, retroviruses like HIV derive from endogenous retroviral elements comprising up to 8% of the human genome, illustrating how genetic parasites can gain extracellular mobility. Critics note that this does not fully explain viruses lacking clear cellular homologs, such as those with unique polymerases.[9][27][28] Conversely, the regressive hypothesis argues that viruses arose from free-living or parasitic cellular organisms, such as ancient bacteria or intracellular symbionts, that progressively lost metabolic genes through reductive evolution while retaining replication machinery dependent on host cells. Evidence includes giant viruses like Mimivirus, discovered in 2003, which encode over 900 genes—including translation components—exceeding those of some unicellular parasites like Rickettsia, and exhibit amoeba host ranges akin to bacterial endosymbionts. This hypothesis aligns with observations of obligate intracellular bacteria undergoing genome shrinkage, from millions to hundreds of base pairs, paralleling viral minimalism. However, it struggles with the antiquity implied, as viral diversification appears to postdate major cellular domains.[9][26][28] The virus-first hypothesis maintains that viruses or virus-like replicators predated or co-evolved alongside the first cells, originating from primordial RNA-protein complexes in a pre-cellular milieu, potentially seeding cellular genomes via gene transfer. Proponents cite the RNA world's reliance on self-replicating ribozymes, mirrored in simple RNA viruses like picornaviruses with genomes under 10 kb, and the ubiquity of viral genes in cellular proteomes—up to 10–20% in some estimates from horizontal transfers. Félix d'Herelle proposed an early version in the 1920s, suggesting viruses as autonomous precursors to cellular life. Challenges include explaining DNA viruses' complexity and the lack of evidence for non-parasitic ancient viruses, as all known viruses require cells for propagation.[9][29][28] Phylogenetic reconstructions indicate polyphyletic origins, with RNA viruses possibly tracing to early RNA-world escapes and DNA viruses to later cellular reductions, reflecting multiple independent emergences rather than a monophyletic event. Ongoing metagenomic surveys, sequencing billions of viral particles from environments like oceans, continue to reveal novel lineages, underscoring the hypotheses' provisional nature amid incomplete sampling of virospheres estimated at 10^31 particles globally.[30][26]Biological Characteristics
Debate on Viral Life Status
The classification of viruses as living or non-living entities remains unresolved in biology, primarily due to ambiguities in defining life itself. Traditional criteria for life, such as those outlined by sources including cellular organization, independent metabolism, homeostasis, growth, and autonomous reproduction, are frequently invoked; viruses fail most of these, as they consist of acellular particles (virions) lacking ribosomes, enzymes for energy production, or self-maintenance capabilities outside a host cell.[31] [32] Extracellular virions exhibit no metabolic activity and cannot replicate without hijacking host cellular machinery, positioning them as inert genetic packages rather than self-sustaining systems.[7] [33] Proponents arguing against viral life status emphasize obligate parasitism: viruses depend entirely on host cells for protein synthesis, energy, and replication, rendering them incapable of independent Darwinian evolution in the strict sense required by definitions like NASA's—a self-sustaining chemical system capable of undergoing evolution—which viruses approximate only through host-mediated processes.[6] This view aligns with the International Committee on Taxonomy of Viruses (ICTV), which treats viruses as distinct from cellular life forms without ascribing vitality to them.[34] Critics of equating viruses with life note that such claims often stem from metaphorical extensions of replication, ignoring the causal dependency on host biochemistry, where the virus directs but does not originate the processes.[35] Conversely, advocates for considering viruses alive highlight their genetic complexity, capacity for mutation and natural selection, and shared molecular building blocks (DNA or RNA encased in protein) with cellular organisms, suggesting they represent a primitive or derived form of life.[36] Some researchers propose the "virocell" concept, wherein an infected host cell transforms into a viral factory, effectively creating a novel living entity with emergent properties like replication and evolvability that transcend the inert virion state.[37] Evolutionary analyses indicate viruses co-evolved with hosts, sharing ancient genes and driving biodiversity, which blurs boundaries and challenges rigid dichotomies.[7] [38] However, these arguments are contested for conflating host-virus dynamics with intrinsic vitality, as the virus alone cannot initiate or sustain these traits without external cellular infrastructure.[39] No scientific consensus exists, with most biologists defaulting to non-living status based on empirical failure to meet core life criteria, though the debate persists due to definitional fluidity and viruses' role in evolution.[40] [41] Virologists like Eugene Koonin argue the question holds little substantive value for research, as viruses function as replicators regardless of categorical labels, prioritizing operational biology over philosophical taxonomy.[42] This ambiguity underscores causal realism in virology: viruses exert profound effects on living systems through genetic hijacking, yet their inert extraceullar form precludes independent agency.[10]Structural Components
Viruses possess a basic structure comprising a nucleic acid genome encased in a protective protein shell known as the capsid, which together form the nucleocapsid core of the virion.[1] The capsid serves to shield the genome from environmental damage and facilitates host cell attachment and entry.[3] In non-enveloped viruses, the capsid constitutes the outermost layer, while enveloped viruses acquire an additional lipid bilayer membrane surrounding the nucleocapsid.[1] The capsid is assembled from repeating structural units called capsomeres, each composed of one or more protein subunits termed protomers.[1] Capsid architectures exhibit three primary symmetries: helical symmetry, seen in elongated viruses such as tobacco mosaic virus where proteins coil around the genome; icosahedral symmetry, approximating a 20-sided polyhedron in spherical viruses like adenoviruses, optimizing enclosure with minimal protein; and complex symmetry, featuring additional structures like tails in bacteriophages such as T4, which include base plates and fibers for host recognition.[1] These symmetries determine virion morphology, with icosahedral capsids often achieving T-numbers (e.g., T=1 for simplest, up to T=25 or higher in larger viruses) that dictate subunit count and stability.[1] Enveloped viruses derive their lipid envelope from modified host cell membranes during budding, incorporating viral glycoproteins that project as spikes for receptor binding and membrane fusion.[1] Underlying the envelope, a matrix protein layer may stabilize the structure and link it to the nucleocapsid, as in influenza viruses where M1 protein regulates assembly.[3] Some viruses include internal components like polymerases within the capsid for immediate replication upon entry, though these are not universal.[1] Virion sizes vary from 20 nm for small RNA viruses like picornaviruses to over 300 nm for complex poxviruses, reflecting structural diversity.[40]Genetic Material and Genome Features
Viruses contain genetic material composed of either DNA or RNA, which serves as the blueprint for viral replication and protein synthesis. This nucleic acid can be single-stranded (ss) or double-stranded (ds), linear or circular, and in some cases segmented into multiple molecules. Unlike cellular organisms, which universally employ dsDNA, viral genomes exhibit this diversity to adapt to host replication machinery and evade defenses.[43][44] Viral genome sizes span several orders of magnitude, from under 3 kilobases (kb) in small ssDNA viruses such as geminiviruses (approximately 2,580 nucleotides) to over 2 megabases (Mb) in giant dsDNA viruses like pandoraviruses (around 2.5 Mb encoding nearly 2,500 proteins). Typical non-giant viral genomes range from 7 to 20 kb, with parvoviruses featuring 4–6 kb ss linear DNA and mimiviruses possessing 1.2 Mb dsDNA. This variation correlates with capsid size and gene count, from a few genes in minimal viruses to hundreds in larger ones, though even giants encode fewer proteins than the smallest cellular genomes.[45][46][1][47][48] Key features of viral genomes include extreme compactness, with high gene density and frequent overlapping genes, where the same nucleotide sequence encodes multiple proteins in different reading frames to maximize coding efficiency within size limits. RNA viruses often produce polyproteins that are cleaved post-translationally, while both DNA and RNA genomes generally lack introns and extensive non-coding regions. Mutation rates are elevated compared to cellular life, particularly in RNA viruses at approximately 10^{-4} substitutions per nucleotide per replication cycle due to error-prone polymerases, fostering rapid evolution; DNA viruses mutate more slowly, leveraging host proofreading enzymes. These traits enable viruses to exploit host resources while maintaining minimal self-sufficiency.[49][50][51][48]Host Dependency and Range
Viruses are obligate intracellular parasites, incapable of independent replication due to the absence of ribosomes, metabolic enzymes, and energy production mechanisms, necessitating hijacking of host cellular machinery for genome duplication, protein synthesis, and virion assembly.[9] This dependency confines viral propagation to viable host cells, where the virus directs host ribosomes to translate viral messenger RNA and commandeers nucleotide pools and polymerases for nucleic acid replication.[52] Outside hosts, viruses persist as inert particles, resistant to environmental stresses but unable to propagate without cellular invasion.[53] The host range of a virus encompasses the spectrum of susceptible species, cell types, or strains it can infect, primarily dictated by molecular compatibility at entry points such as receptor-binding specificity of viral surface proteins to host glycoproteins or receptors.[54] Intracellular barriers further restrict range, including uncoating efficiency, exploitation of host replication factors, and circumvention of antiviral defenses like interferon responses or restriction factors.[55] For influenza A viruses, host specificity arises from hemagglutinin adaptations to sialic acid receptor linkages varying across species—alpha-2,6 in humans versus alpha-2,3 in birds—enabling avian strains to occasionally adapt for mammalian transmission.[56] Viruses display diverse host ranges, from highly narrow to exceptionally broad, influencing transmission dynamics and zoonotic potential. Narrow-range examples include infectious hematopoietic necrosis virus (IHNV), confined to salmonid fish species, reflecting stringent receptor and replication compatibilities.[57] Conversely, cucumber mosaic virus infects over 1,200 plant species across multiple families, underscoring broad receptor promiscuity and metabolic adaptability in botanical hosts.[58] SARS-CoV-2 demonstrates intermediate breadth, productively infecting primates, felines, mustelids, and suids via angiotensin-converting enzyme 2 (ACE2) receptor variations, though inefficiently in rodents without adaptation.[59] Bacteriophages typically exhibit strain-specific narrow ranges within bacterial genera, though selection on diverse hosts can yield broader variants capable of lysing multiple strains.[60] Host range expansions often occur via mutations altering attachment proteins or recombination with co-infecting viruses, as seen in historical influenza pandemics originating from avian reservoirs adapting to human cells.[56] Such shifts underscore causal links between ecological interfaces—like wildlife-livestock interfaces—and emergence risks, with empirical surveillance data revealing most human viruses derive from animal origins.[57] Tissue tropism within permissive hosts adds granularity, where viruses like poliovirus target neuronal subsets via specific receptor distributions, amplifying pathogenicity despite broader cellular susceptibility in vitro.[61]Replication and Dynamics
Infection Mechanisms
Viruses initiate infection by attaching to specific receptors on the host cell surface, a process mediated by interactions between viral surface proteins—such as glycoproteins on enveloped viruses or capsid proteins on non-enveloped viruses—and host cell molecules including proteins, carbohydrates, or lipids.[62] This receptor binding determines viral host range and tropism, as the availability of compatible receptors dictates susceptible cell types; for instance, human immunodeficiency virus (HIV) targets CD4 receptors on T cells, while influenza A virus binds sialic acid residues.[63] Attachment often induces conformational changes in viral proteins, priming the virion for subsequent entry steps, and may involve co-receptors or accessory molecules to enhance specificity and efficiency.[62] Following attachment, viral entry occurs via distinct pathways tailored to the virus structure and host. Enveloped viruses frequently employ direct membrane fusion at the plasma membrane, where viral fusion proteins (e.g., hemagglutinin in influenza) undergo pH-independent or receptor-triggered rearrangements to merge the viral envelope with the host lipid bilayer, releasing the capsid into the cytoplasm.[64] Alternatively, many enveloped viruses, such as HIV and Ebola, enter through endocytosis, where receptor-bound virions are internalized into endosomes; low pH or proteolytic cleavage in the endosome then activates fusion proteins to breach the endosomal membrane.[65] Non-enveloped viruses, lacking a lipid envelope, typically rely on endocytic uptake—via clathrin-coated pits, caveolae, or macropinocytosis—followed by endosomal escape through capsid disassembly, pore formation, or membrane penetration by viral peptides, as seen in adenoviruses utilizing protein VI for lysis.[66] Bacteriophages, which infect bacterial hosts, often bypass endocytosis altogether, injecting nucleic acid directly through the cell wall and membrane via a tail fiber-mediated attachment and syringe-like apparatus, as exemplified by T4 phage piercing peptidoglycan layers.[67] These mechanisms exploit host cellular machinery while evading innate defenses like phagocytosis or receptor downregulation, though efficiency varies; for example, receptor density influences infection rates, with low-affinity bindings requiring higher viral titers.[68] Entry failures, due to mismatched receptors or host restrictions, underpin viral specificity and inform antiviral strategies targeting these initial steps.[69]Replication Cycle
Viruses, lacking independent metabolic machinery, replicate solely within host cells by commandeering cellular ribosomes, enzymes, and energy sources for genome duplication and protein synthesis.[8] The standard replication cycle encompasses attachment, penetration, uncoating, replication, assembly, maturation, and release, though specifics vary by viral family.[70][71] Attachment initiates the cycle as viral capsid or envelope glycoproteins bind to specific host cell surface receptors, conferring tropism and host range.[71] Penetration ensues via receptor-mediated endocytosis, direct membrane fusion for enveloped viruses, or nucleic acid injection as in bacteriophages.[70] Uncoating follows, wherein host or viral proteases dismantle the capsid, liberating the genome into the cytoplasm or nucleus.[71] Replication phase exploits host machinery: DNA viruses typically transcribe mRNA and replicate in the nucleus using cellular polymerases, while RNA viruses often operate cytoplasmically, with positive-sense RNA serving directly as mRNA and negative-sense requiring viral RNA-dependent RNA polymerase.[8] Retroviruses like HIV reverse-transcribe RNA to DNA for nuclear integration.[8] Viral proteins, including structural components, are translated on host ribosomes.[71] Assembly packages replicated genomes with synthesized proteins into progeny virions, often self-assembling spontaneously; maturation may involve proteolytic cleavage for infectivity.[8] Release disperses virions: non-enveloped viruses via host cell lysis in the lytic cycle, or enveloped ones through budding, acquiring lipid envelopes from host membranes without immediate lysis.[71][70] Bacteriophages exhibit lytic cycles culminating in host lysis or lysogenic cycles integrating phage DNA as prophage into bacterial genomes, replicating passively during cell division until environmental cues trigger lytic induction.[72] Analogous latency occurs in eukaryotic viruses like herpesviruses, evading immunity via genome persistence without active production.[8]Genetic Variation and Evolution
Viruses exhibit high rates of genetic variation due to error-prone replication mechanisms, enabling rapid evolution in response to selective pressures such as host immunity and antiviral therapies. Mutation rates in RNA viruses typically range from 10^{-6} to 10^{-4} substitutions per nucleotide per replication cycle, orders of magnitude higher than the 10^{-8} to 10^{-6} observed in DNA viruses, primarily because most RNA-dependent RNA polymerases lack 3'–5' exonuclease proofreading activity.[73][74] DNA viruses, by contrast, often benefit from host cellular proofreading enzymes during replication, resulting in lower fidelity errors.[75] These elevated mutation frequencies, combined with short generation times and large population sizes within hosts, generate substantial genetic diversity per infection cycle.[76] Beyond point mutations, viruses diversify through recombination—exchange of genetic segments between co-infecting strains—and reassortment in segmented genomes, where entire gene segments are swapped.[77] Recombination occurs via template switching during replication or breakage and rejoining of nucleic acids, prevalent in RNA viruses like coronaviruses and retroviruses.[78] Reassortment, unique to viruses with multipartite genomes such as influenza, allows rapid assembly of novel combinations from parental strains in doubly infected cells, facilitating jumps to new hosts or evasion of immunity.[79] These processes amplify variation beyond mutation alone, with natural selection favoring fit variants amid genetic drift and bottlenecks during transmission.[80] The quasispecies model describes viral populations as dynamic clouds of closely related mutants rather than uniform clones, arising from mutation rates exceeding the error threshold for consensus sequence maintenance.[81] In this framework, intra-host diversity enables collective adaptation, with defective genomes potentially complementing fit ones, though selection prunes deleterious variants over time.[82] RNA viruses like poliovirus exemplify this, maintaining mutant swarms that enhance adaptability to environmental stresses.[83] Evolutionary rates vary widely, with substitution rates spanning six orders of magnitude influenced by genomic architecture, replication speed, and selection intensity rather than mutation rate alone.[80] For instance, HIV-1, an RNA retrovirus, accumulates substitutions at approximately 10^{-3} per site per year, driven by reverse transcriptase errors and recombination, enabling immune escape and drug resistance.[84] Influenza A undergoes antigenic drift via gradual mutations in hemagglutinin and neuraminidase genes, necessitating annual vaccine updates, while major shifts from reassortment caused pandemics like 1957 (H2N2) and 1968 (H3N2).[85] SARS-CoV-2 evolves more slowly than HIV or influenza, with a rate of about 10^{-3} substitutions per site per year, yet variants like Omicron (B.1.1.529, detected November 2021) emerged through recombination and selection for transmissibility and immune evasion.[86][87] These patterns underscore viruses' capacity for adaptive evolution, constrained by host factors and transmission dynamics.[88]Persistent and Latent Infections
Persistent infections occur when a virus establishes long-term presence in the host without complete clearance, often involving continuous or intermittent replication at low levels that avoids rapid host cell destruction or immune elimination.[89] These infections contrast with acute ones by persisting beyond the initial symptomatic phase, typically through mechanisms such as antigenic variation, downregulation of viral gene expression, or integration into the host genome, allowing the virus to evade adaptive immunity while maintaining infectivity in reservoirs like lymphocytes or epithelial cells.[89] Host factors, including immune suppression or genetic predispositions, contribute to persistence, as seen in cases where defective immune responses fail to eliminate infected cells.[90] Latent infections represent a subset of persistent infections characterized by the viral genome's dormancy within host cells, with minimal or no viral protein production and no active replication, enabling indefinite survival without immediate pathogenesis.[91] Latency is maintained by epigenetic silencing of viral promoters, such as histone modifications or microRNA interference, which repress lytic genes while preserving the genome in episomal or integrated forms, often in non-dividing cells like neurons.[92] Reactivation can be triggered by stressors like immunosuppression, hormonal changes, or UV exposure, shifting to productive replication and shedding.[93] This strategy ensures viral propagation through asymptomatic carriers, as the latent phase imposes no fitness cost on the host until reactivation.[94] Mechanisms distinguishing persistent from strictly latent infections include ongoing low-level virion production in the former, which sustains chronic inflammation or immune exhaustion, versus transcriptional quiescence in latency that permits immune tolerance without constant antigen exposure.[95] For instance, in persistent infections like hepatitis B virus (HBV), the virus replicates in hepatocytes via reverse transcription of pregenomic RNA, leading to circulating Dane particles and immune complex-mediated liver damage over decades.[89] In contrast, herpes simplex virus type 1 (HSV-1) establishes latency in trigeminal ganglia, where the genome circularizes as an episome with only latency-associated transcripts (LATs) expressed, suppressing immediate-early genes until neuronal stress induces lytic reactivation, causing recurrent oral lesions in approximately 20-40% of carriers annually.[96] Human immunodeficiency virus (HIV) exemplifies a hybrid persistent infection with latent reservoirs in resting CD4+ T cells, where integrated proviral DNA remains transcriptionally silent due to chromatin compaction, resisting antiretroviral therapy and contributing to rebound upon treatment cessation.[90] These dynamics highlight how viruses exploit host cellular machinery for survival, often leading to lifelong carriage and potential oncogenic risks, as in Epstein-Barr virus latency linked to Burkitt's lymphoma via restricted gene expression in B lymphocytes.[96] Persistent and latent infections pose challenges for eradication, as antiviral drugs targeting replication fail against dormant genomes, necessitating strategies like latency-reversing agents to expose hidden reservoirs for immune clearance.[92] Examples include varicella-zoster virus (VZV), which after primary chickenpox integrates latently in dorsal root ganglia and reactivates as shingles in 30% of infected individuals over age 80 due to waning cell-mediated immunity.[96] Such infections underscore viral adaptation to host longevity, prioritizing transmission over virulence in stable populations.[90]Classification
ICTV Taxonomic Framework
The International Committee on Taxonomy of Viruses (ICTV), established in 1971 under the International Union of Microbiological Societies, serves as the authoritative body for naming and classifying viruses and virus-like entities worldwide.[97] It maintains a universal taxonomy based on shared properties, evolutionary relationships, and genetic characteristics, distinguishing viruses as physical entities from the abstract taxa to which they are assigned.[98] This framework excludes viruses from the three-domain system of cellular life (Bacteria, Archaea, Eukarya), instead employing a dedicated hierarchy that begins at the rank of realm to accommodate viral diversity.[99] The ICTV taxonomy utilizes up to 15 hierarchical ranks, expanded from five traditional levels (order, family, subfamily, genus, species) in 2020 to better align with Linnaean principles while partitioning the virosphere into discrete groups.[100] These ranks, in descending order of inclusivity, are: realm, subrealm, kingdom, subkingdom, phylum, subphylum, class, subclass, order, suborder, family, subfamily, genus, subgenus, and species.[99] Classification decisions are guided by empirical data on virion morphology, genome type and organization, replication strategy, and phylogenetic analyses, with proposals ratified by ICTV executive and study group votes.[97] As of the August 2025 release (following February 2025 ratifications under Master Species List #40), the taxonomy encompasses 7 realms, 11 kingdoms, 22 phyla, 4 subphyla, 49 classes, 93 orders, 12 suborders, over 800 families, and thousands of species, reflecting ongoing discoveries in viral metagenomics.[101] Key realms include Adnaviria (featuring linear double-stranded DNA in A-form conformation, infecting archaea and bacteria), Duplodnaviria (double-stranded DNA viruses with diverse hosts, including bacteriophages), Monodnaviria (monopartite single-stranded DNA viruses), Riboviria (RNA viruses using RNA-dependent RNA polymerase), Ribozyviria (ribozyme-based replication in viroids and relatives), Shotokuvirae (segmented RNA viruses), and Varidnaviria (vertically transmitted DNA viruses with icosahedral symmetry).[102][103] These higher ranks group viruses by fundamental replication mechanisms and genome architectures, with lower ranks refining based on host range, protein homology, and sequence divergence.[100] Recent updates, such as standardized binomial species names (genus epithet plus descriptive term), enhance precision and interoperability with genomic databases.[104] The framework evolves through proposal-based amendments, prioritizing monophyly and diagnostic traits over host specificity alone, though debates persist on accommodating uncultured viruses identified via metagenomics.[105] ICTV resources, including the online taxonomy browser and annual reports, provide searchable access to ratified classifications, ensuring the system's adaptability to new data without retroactive overhauls.[101] This structure underpins virological research by offering a stable, evidence-driven nomenclature that facilitates cross-study comparisons.[106]Baltimore Classification System
The Baltimore classification system, proposed by virologist David Baltimore in 1971, divides viruses into seven groups based on the type of nucleic acid in their genome and the specific pathway used to synthesize messenger RNA (mRNA) from that genome.[107] This functional scheme focuses on the central dogma of molecular biology—particularly the flow of genetic information from genome to mRNA—rather than morphological or host-range criteria, providing a framework that highlights diverse replication strategies among viruses.[108] Baltimore's original publication outlined six classes, with the seventh added later to accommodate viruses that use reverse transcriptase despite possessing a partially double-stranded DNA genome.[107] Viruses in Group I contain double-stranded DNA (dsDNA) genomes and produce mRNA via host RNA polymerase transcription, similar to cellular DNA. Examples include adenoviruses, herpesviruses, and poxviruses.[109] Group II viruses have single-stranded DNA (ssDNA) genomes, which are converted to dsDNA intermediates before transcription to mRNA; parvoviruses exemplify this group.[109] Group III features double-stranded RNA (dsRNA) genomes, where viral RNA-dependent RNA polymerase transcribes mRNA from one strand; reoviruses are representative.[109] Group IV viruses possess positive-sense single-stranded RNA (+ssRNA) genomes that function directly as mRNA upon entry into the host cell, enabling immediate translation; picornaviruses and coronaviruses belong here.[110] In contrast, Group V viruses have negative-sense single-stranded RNA (-ssRNA) genomes requiring transcription by viral RNA-dependent RNA polymerase to generate +mRNA; examples include rhabdoviruses (e.g., rabies virus) and paramyxoviruses (e.g., measles virus).[109] Group VI comprises retroviruses with +ssRNA genomes that are reverse-transcribed into DNA, which integrates into the host genome for subsequent mRNA production; human immunodeficiency virus (HIV) is a key example.[110] Group VII includes viruses with partially dsDNA genomes that replicate via an RNA intermediate using reverse transcriptase, such as hepadnaviruses like hepatitis B virus.[107] This classification remains influential for understanding viral gene expression and has informed antiviral drug development by targeting group-specific enzymes, like reverse transcriptase inhibitors for Groups VI and VII.[108] While orthogonal to the International Committee on Taxonomy of Viruses (ICTV) hierarchy, it complements taxonomic efforts by emphasizing mechanistic differences.[111]| Group | Genome Type | mRNA Synthesis Mechanism | Examples |
|---|---|---|---|
| I | dsDNA | Direct transcription by host RNA polymerase | Adenoviruses, herpesviruses, poxviruses[109] |
| II | ssDNA | Conversion to dsDNA, then transcription | Parvoviruses[109] |
| III | dsRNA | Transcription by viral RdRp from dsRNA template | Reoviruses[109] |
| IV | +ssRNA | Genome serves as mRNA | Picornaviruses, coronaviruses[110] |
| V | -ssRNA | Transcription by viral RdRp to +mRNA | Rhabdoviruses, paramyxoviruses[109] |
| VI | +ssRNA-RT | Reverse transcription to DNA, integration, then transcription | Retroviruses (e.g., HIV)[110] |
| VII | dsDNA-RT | RNA intermediate via reverse transcription for replication | Hepadnaviruses (e.g., hepatitis B)[107] |