Recent from talks
All channels
Be the first to start a discussion here.
Be the first to start a discussion here.
Be the first to start a discussion here.
Be the first to start a discussion here.
Welcome to the community hub built to collect knowledge and have discussions related to Glossary of cellular and molecular biology (M–Z).
Nothing was collected or created yet.
Glossary of cellular and molecular biology (M–Z)
View on Wikipediafrom Wikipedia
This glossary of cellular and molecular biology is a list of definitions of terms and concepts commonly used in the study of cell biology, molecular biology, and related disciplines, including molecular genetics, biochemistry, and microbiology.[1] It is split across two articles:
- Glossary of cellular and molecular biology (0–L) lists terms beginning with numbers and those beginning with the letters A through L.
- Glossary of cellular and molecular biology (M–Z) (this page) lists terms beginning with the letters M through Z.
This glossary is intended as introductory material for novices (for more specific and technical detail, see the article corresponding to each term). It has been designed as a companion to Glossary of genetics and evolutionary biology, which contains many overlapping and related terms; other related glossaries include Glossary of virology and Glossary of chemistry.
M
[edit]- M phase
- See mitosis.
- macromolecule
- Any very large molecule composed of dozens, hundreds, or thousands of covalently bonded atoms, especially one with biological significance. Many important biomolecules, such as nucleic acids and proteins, are polymers consisting of a repeated series of smaller monomers; others such as lipids and carbohydrates may not be polymeric but are nevertheless large and complex molecules.
- macronucleus
- The larger of the two types of nuclei which occur in pairs in the cells of some ciliated protozoa. Macronuclei are highly polyploid and responsible for directing vegetative reproduction, in contrast to the diploid micronuclei, which have important functions during conjugation.[2]
- macrophage
- Any of a class of relatively long-lived phagocytic cells of the mammalian immune system which are activated in response to the presence of foreign materials in certain tissues and subsequently play important roles in antigen presentation, stimulating other types of immune cells, and killing or engulfing parasitic microorganisms, diseased cells, or tumor cells.[3]
- major groove
- map-based cloning
- See positional cloning.
- massively parallel sequencing
- medical genetics
- The branch of medicine and medical science that involves the study, diagnosis, and management of hereditary disorders, and more broadly the application of knowledge about human genetics to medical care.
- megabase (Mb)
- A unit of nucleic acid length equal to one million (1×106) bases in single-stranded molecules or one million base pairs in duplex molecules such as double-stranded DNA.
- meiosis
- A specialized type of cell division that occurs exclusively in sexually reproducing eukaryotes, during which DNA replication is followed by two consecutive rounds of division to ultimately produce four genetically unique haploid daughter cells, each with half the number of chromosomes as the original diploid parent cell. Meiosis only occurs in cells of the sex organs, and serves the purpose of generating haploid gametes such as sperm, eggs, or spores, which are later fused during fertilization. The two meiotic divisions, known as Meiosis I and Meiosis II, may also include various genetic recombination events between homologous chromosomes.
- meiotic spindle
- See spindle apparatus.
- melting
- The denaturation of a double-stranded nucleic acid into two single strands, especially in the context of the polymerase chain reaction.
- membrane
- A supramolecular aggregate of amphipathic lipid molecules which when suspended in a polar solvent tend to arrange themselves into structures which minimize the exposure of their hydrophobic tails by sheltering them within a ball created by their own hydrophilic heads (i.e. a micelle). Certain types of lipids, specifically phospholipids and other membrane lipids, commonly occur as double-layered sheets of molecules when immersed in an aqueous environment, which can themselves assume approximately spherical shapes, acting as semipermeable barriers surrounding a water-filled interior space. This is the basic structure of the biological membranes enclosing all cells, vesicles, and membrane-bound organelles.
- membrane lipid
- membrane potential
- membrane protein
- Any protein that is closely associated either transiently or permanently with the lipid bilayer membrane surrounding a cell, organelle, or vesicle.[4]
- membrane-bound organelle
- An organelle or cellular compartment enclosed by its own dedicated lipid membrane, separating its interior from the rest of the cytoplasm.
- messenger RNA (mRNA)
- Any of a class of single-stranded RNA molecules which function as molecular messengers, carrying sequence information encoded in the DNA genome to the ribosomes where protein synthesis occurs. The primary products of transcription, mRNAs are synthesized by RNA polymerase, which builds a chain of ribonucleotides that complement the deoxyribonucleotides of a DNA template; in this way, the DNA sequence of a protein-coding gene is effectively preserved in the raw transcript, which is subsequently processed into a mature mRNA by a series of post-transcriptional modifications.
- metabolic pathway
- A stepwise series of biochemical reactions occurring within a cell, often but not necessarily catalyzed by specific enzymes, that fulfills some activity or process related to metabolism. The reactions are linked by the sharing of reactants, products, or intermediate compounds in consecutive steps, such that the product of one reaction is used as a reactant in a subsequent reaction. Byproducts are often removed from the cell as metabolic waste. The overall pathway may be anabolic, catabolic, or amphibolic in nature.[5] In any actively metabolizing cell, an elaborate network of interconnected metabolic pathways is required to maintain homeostasis, with degradative catabolic processes providing the energy necessary to conduct anabolic biosynthesis; for example, glycolysis, the electron transport chain, and oxidative phosphorylation provide the ATP used in fatty acid synthesis. The flux of metabolites through each pathway is regulated by the needs of the cell and the availability of substrates.
- metabolic waste
- metabolism
- The complete set of chemical reactions which sustain and account for the basic processes of life in all living cells,[2] especially those involving: 1) the conversion of energy from food into energy available for cellular activities; 2) the breakdown of food into simpler compounds which can then be used as substrates to build complex biomolecules such as proteins, lipids, and nucleic acids; and 3) the degradation and excretion of toxins, byproducts, and other unusable compounds known as metabolic wastes. In a broader sense the term may include all chemical reactions occurring in living organisms, even those which are not strictly necessary for life but instead serve accessory functions. Many specific cellular activities are accomplished by metabolic pathways in which one chemical is ultimately transformed through a stepwise series of reactions into another chemical, with each reaction catalyzed by a specific enzyme. Most metabolic reactions can be subclassified as catabolic or anabolic.
- metabolite
- An intermediate or end product of metabolism, especially degradative metabolism (catabolism);[2] or any substance produced by or taking part in a metabolic reaction. Metabolites include a huge variety of small molecules generated by cells from various pathways and having various functions, including as inputs to other pathways and reactions, as signaling molecules, and as stimulators, inhibitors, and cofactors of enzymes. Metabolites may result from the degradation and elimination of naturally occurring compounds as well as of synthetic compounds such as pharmaceuticals.
- metabolome
- The complete set of small-molecule chemical compounds within a cell, organelle, or any other biological sample, including both endogenous molecules (e.g. individual amino acids and nucleotides, fatty acids, organic acids, amines, simple sugars, vitamins, antibiotics, etc.) and exogenous molecules (e.g. drugs, toxins, environmental contaminants, and other xenobiotics).
- metacentric
- (of a linear chromosome or chromosome fragment) Having a centromere positioned in the middle of the chromosome, resulting in chromatid arms of approximately equal length.[6]
- metaphase
- The stage of mitosis and meiosis that occurs after prometaphase and before anaphase, during which the centromeres of the replicated chromosomes align along the equator of the cell, with each kinetochore attached to the mitotic spindle.
- methylation
- The covalent attachment of a methyl group (–CH
3) to a chemical compound, protein, or other biomolecule, either spontaneously or by enzymatic catalysis. Methylation is one of the most widespread natural mechanisms by which nucleic acids and proteins are labelled. The methylation of nucleobases in a DNA molecule inhibits recognition of the methylated sequence by DNA-binding proteins, which can effectively silence the expression of genes. Specific residues within histones are also commonly methylated, which can change nucleosome positioning and similarly activate or repress nearby loci. The opposite reaction is demethylation. - methyltransferase
- Any of a class of transferase enzymes which catalyze the covalent bonding of a methyl group (–CH
3) to another compound, protein, or biomolecule, a process known as methylation. - MicroArray and Gene Expression (MAGE)
- A group that "aims to provide a standard for the representation of DNA microarray gene expression data that would facilitate the exchange of microarray information between different data systems".[7]
- microbody
- Any of a diverse class of small membrane-bound organelles or vesicles found in the cells of many eukaryotes, especially plants and animals, usually having some specific metabolic function and occurring in great numbers in certain specialized cell types. Peroxisomes, glyoxysomes, glycosomes, and hydrogenosomes are often considered microbodies.
- microchromosome
- A type of very small chromosome, generally less than 20,000 base pairs in size, present in the karyotypes of some organisms.
- microdeletion
- A chromosomal deletion that is too short to cause any apparent change in morphology under a light microscope, though it may still be detectable with other methods such as sequencing.
- microfilament
- A long, thin, flexible, rod-like structure composed of polymeric strands of proteins, usually actins, that occurs in abundance in the cytoplasm of eukaryotic cells, forming part of the cytoskeleton. Microfilaments comprise the cell's structural framework. They are modified by and interact with numerous other cytoplasmic proteins, playing important roles in cell stability, motility, contractility, and facilitating changes in cell shape, as well as in cytokinesis.
- micronucleus
- The smaller of the two types of nuclei that occur in pairs in the cells of some ciliated protozoa. Whereas the larger macronucleus is polyploid, the micronucleus is diploid and generally transcriptionally inactive except for the purpose of sexual reproduction, where it has important functions during conjugation.[2]
- microRNA (miRNA)
- A type of small, single-stranded, non-coding RNA molecule that functions in post-transcriptional regulation of gene expression, particularly RNA silencing, by base-pairing with complementary sequences in mRNA transcripts, which typically results in the cleavage or destabilization of the transcript or inhibits its translation by ribosomes.
- microsatellite
- A type of satellite DNA consisting of a relatively short sequence of tandem repeats, in which certain motifs (ranging in length from one to six or more bases) are repeated, typically 5–50 times. Microsatellites are widespread throughout most organisms' genomes and tend to have higher mutation rates than other regions. They are classified as variable number tandem repeat (VNTR) DNA, along with longer minisatellites.
- microsome
- A small intracellular vesicle derived from fragments of endoplasmic reticulum observed in cells which have been homogenized.[4]
- microspike
- See filopodium.
- microtome
- An instrument used to cut extremely thin slices of material, known as microsections or simply sections, preparatory to observation under a microscope.[5] Sections of tissues and cells are usually 50 nanometres (nm) to 100 micrometres (μm) in width. The process of cutting them is known as microtomy.
- microtrabecula
- A fine protein filament of the cytoskeleton. Multiple filaments form the microtrabecular network.[2]
- microtubule
- Any of the long, generally straight, hollow tubes, about 24 nanometers in diameter and composed of interwoven polymeric filaments of the protein tubulin, found in the cytoplasm of many eukaryotic cells, where they are involved in maintaining the cell's shape and structural integrity as well as in force generation for cellular or organellar locomotion (as with cilia and flagella). They also comprise the spindle apparatus critical to mitosis and meiosis. Microtubules are rigid but transient all-purpose structural members which can be rapidly assembled and disassembled at the cell's needs. Many different microtubule-associated proteins interact with them.[5] See also microfilament.
- microtubule-organizing center (MTOC)
- A region near the center of a eukaryotic cell typically consisting of two centrioles oriented at right angles to each other and surrounded by a complex of associated proteins, which functions as the site of initiation for the assembly of microtubules.[5]
- microvesicle
- A type of extracellular vesicle released when an evagination of the cell membrane "buds off" into the extracellular space. Microvesicles vary in size from 30–1,000 nanometres in diameter and are thought to play roles in many physiological processes, including intercellular communication by shuttling molecules such as RNA and proteins between cells.[8]
- microvillus
- A small, slender, tubular cytoplasmic projection, generally 0.2–4 micrometres long and 0.1 micrometres in diameter,[9] protruding from the surface of some animal cells and supported by a central core of microfilaments. When present in large numbers, such as on epithelial cells lining the respiratory and alimentary tracts, they form a dense brush border which presumably serves to increase each cell's absorptive surface area.[2][3]
- mid body
- The centrally constricted region that forms across the central axis of a cell during cytokinesis, constricted by the closing of the contractile ring until the daughter cells are finally separated,[2] but occasionally persisting as a tether between the two cells for as long as a complete cell cycle.[9]
- middle lamella
- In plant cells, the outermost layer of the cell wall; a continuous, unified layer of extracellular pectins which is the first layer deposited by the cell during cytokinesis and which serves to cement together the primary cell walls of adjacent cells.[4]
- Minimal information about a high-throughput sequencing experiment (MINSEQE)
- A commercial standard developed by FGED for the storage and sharing of high-throughput sequencing data.[10]
- Minimum information about a microarray experiment (MIAME)
- A commercial standard developed by FGED and based on MAGE in order to facilitate the storage and sharing of gene expression data.[11][12]
- minisatellite
- A region of repetitive, non-coding genomic DNA in which certain DNA motifs (typically 10–60 bases in length) are tandemly repeated (typically 5–50 times). In the human genome, minisatellites occur at more than 1,000 loci, especially in centromeres and telomeres, and exhibit high mutation rates and high variability between individuals. Like the shorter microsatellites, they are classified as variable number tandem repeats (VNTRs) and are a type of satellite DNA.
- minor groove
- minus-strand
- See template strand.
- miRNA
- See microRNA.
- mismatch
- An incorrect pairing of nucleobases on complementary strands of DNA or RNA; i.e. the presence in one strand of a duplex molecule of a base that is not complementary (by Watson–Crick pairing rules) to the base occupying the corresponding position in the other strand, which prevents normal hydrogen bonding between the bases. For example, a guanine paired with a thymine would be a mismatch, as guanine normally pairs with cytosine.[13]
- mismatch repair (MMR)
- missense mutation
- A type of point mutation which results in a codon that codes for a different amino acid than in the unmutated sequence. Compare nonsense mutation.
- mistranslation
- The insertion of an incorrect amino acid in a growing peptide chain during translation, i.e. the inclusion of any amino acid that is not the one specified by a particular codon in an mRNA transcript. Mistranslation may originate from a mischarged transfer RNA or from a malfunctioning ribosome.[13]
- mitochondrial DNA (mtDNA)
- The set of DNA molecules contained within mitochondria, usually one or more circular plasmids representing a semi-autonomous genome which is physically separate from and functionally independent of the chromosomal DNA in the cell's nucleus. The mitochondrial genome encodes many unique enzymes found only in mitochondria.
- mitochondrial fusion
- mitochondrion
- A highly pleiomorphic membrane-bound organelle found in the cytoplasm of nearly all eukaryotic cells, usually in large numbers in the form of sausage-shaped structures 5–10 micrometres in length,[9] enclosed by a double membrane, with the inner membrane infolded in an elaborate series of cristae so as to maximize surface area. Mitochondria are the primary sites of ATP synthesis, where ATP is regenerated from ADP via oxidative phosphorylation, as well as many supporting pathways, including the citric acid cycle and the electron transport chain.[3] Like other plastids, mitochondria contain their own genome encoded in circular DNA molecules which replicate independently of the nuclear genome, as well as their own unique set of transcription factors, polymerases, ribosomes, transfer RNAs, and aminoacyl-tRNA synthetases with which to direct transcription and translation of their genes. The majority of the structural proteins found in mitochondria are encoded by nuclear genes, however, such that mitochondria are only partially autonomous.[2] These observations suggest mitochondria evolved from symbiotic prokaryotes living inside eukaryotic cells.
- mitogen
- Any substance or stimulus that promotes or induces mitosis, or more generally which causes cells to re-enter the cell cycle.[3]
- mitophagy
- The selective degradation of mitochondria by means of autophagy; i.e. the mitochondrion initiates its own degradation. Mitophagy is a regular process in healthy populations of cells by which defective or damaged mitochondria are recycled, preventing their accumulation. It may also occur in response to the changing metabolic needs of the cell, e.g. during certain developmental stages.
- mitosis
- In eukaryotic cells, the part of the cell cycle during which the division of the nucleus takes place and replicated chromosomes are separated into two distinct nuclei. Mitosis is generally preceded by the S phase of interphase, when the cell's DNA is replicated, and either occurs simultaneously with or is followed by cytokinesis, when the cytoplasm and plasma membrane are divided into two new daughter cells. Colloquially, the term "mitosis" is often used to refer to the entire process of cell division, not just the division of the nucleus.
- mitotic index (MI)
- The proportion of cells within a sample which are undergoing mitosis at the time of observation, typically expressed as a percentage or as a value between 0 and 1. The number of cells dividing by mitosis at any given time can vary widely depending on organism, tissue, developmental stage, and culture media, among other factors.[2]
- mitotic recombination
- The abnormal exchange of genetic material between homologous chromosomes during mitosis (as opposed to meiosis, where it occurs normally). Homologous recombination during mitosis is relatively uncommon; in the laboratory, it can be induced by exposing dividing cells to high-energy electromagnetic radiation such as X rays. As in meiosis, it can separate heterozygous alleles and thereby propagate potentially significant changes in zygosity to daughter cells, though unless it occurs very early in development this often has little or no phenotypic effect, since any phenotypic variance shown by mutant lineages arising in terminally differentiated cells is generally masked or compensated for by neighboring wild-type cells.[2]
- mitotic rounding
- The process by which most animal cells undergo an overall change in shape during or preceding mitosis, abandoning the various complex or elongated shapes characteristic of interphase and rapidly contracting into a rounded or spherical morphology that is more conducive to cell division. This phenomenon has been observed both in vivo and in vitro.
- mitotic segregation
- mitotic spindle
- See spindle apparatus.
- mixoploidy
- The presence of more than one different ploidy level, i.e. more than one number of sets of chromosomes, in different cells of the same cellular population.[13]
- mobile genetic element (MGE)
- Any genetic material that can move between different parts of a genome or be transferred from one species or replicon to another within a single generation. The many types of MGEs include transposable elements, bacterial plasmids, bacteriophage elements which integrate into host genomes by viral transduction, and self-splicing introns.
- mobilome
- The complete set of mobile genetic elements within a particular genome, cell, species, or other taxon, including all transposons, plasmids, prophages, and other self-splicing nucleic acid molecules.
- molecular biology
- The branch of biology that studies biological activity at the molecular level, in particular the various mechanisms underlying the biological processes that occur in and between cells, including the structures, properties, synthesis, and modification of biomolecules such as proteins and nucleic acids, their interactions with the chemical environment and with other biomolecules, and how these interactions explain the observations of classical biology (which in contrast studies biological systems at much larger scales).[14] Molecular biology relies largely on laboratory techniques of physics and chemistry to manipulate and measure microscopic phenomena. It is closely related to and overlaps with the fields of cell biology, biochemistry, and molecular genetics.
- molecular cloning
- Any of various molecular biology methods designed to replicate a particular molecule, usually a DNA sequence or a protein, many times inside the cells of a natural host. Commonly, a recombinant DNA fragment containing a gene of interest is ligated into a plasmid vector, which competent bacterial cells are then induced to uptake in a process known as transformation. The bacteria, carrying the recombinant plasmid, are then allowed to proliferate naturally in cell culture, so that each time the bacterial cells divide, the plasmids are replicated along with the rest of the bacterial genome. Any functioning gene of interest within the plasmid will be expressed by the bacterial cells, and thereby its gene products will also be cloned. The plasmids or gene products, which now exist in many copies, may then be extracted from the bacteria and purified. Molecular cloning is a fundamental tool of genetic engineering employed for a wide variety of purposes, often to study gene expression, to amplify a specific gene product, or to generate a selectable phenotype.
- molecular genetics
- A branch of genetics that employs methods and techniques of molecular biology to study the structure and function of genes and gene products at the molecular level. Contrast classical genetics.
- monad
- A haploid set of chromosomes as it exists inside the nucleus of an immature gametic cell such as an ootid or spermatid, i.e. a cell which is a product of meiosis but is not yet a mature gamete.[13]
- monocentric
- (of a linear chromosome or chromosome fragment) Having only one centromere. Contrast dicentric and holocentric.
- monoclonal
- Describing cells, proteins, or molecules descended or derived from a single clone (i.e. from the same genome or genetic lineage) or made in response to a single unique compound. Monoclonal antibodies are raised against only one antigen or can only recognize one unique epitope on the same antigen. Similarly, the cells of some tissues and neoplasms may be described as monoclonal if they are all the asexual progeny of one original parent cell.[2] Contrast polyclonal.
- monocyte
- A type of large leukocyte of the mononuclear phagocyte system in mammals, characterized by pale-staining cytoplasm and a kidney-shaped or horseshoe-shaped nucleus. Monocytes are derived from pluripotent stem cells in bone marrow and become macrophages in other tissues.[5]
- monokaryotic
- (of a cell) Having a single nucleus, as opposed to no nucleus or multiple nuclei.
- monomer
- A molecule or compound which can exist individually or serve as a building block or subunit of a larger macromolecular aggregate known as a polymer.[4] Polymers form when multiple monomers of the same or similar molecular species are connected to each other by chemical bonds, either in a linear chain or a non-linear conglomeration. Examples include the individual nucleotides which form nucleic acid polymers, the individual amino acids which form polypeptides, and the individual proteins which form protein complexes.
- monoploid
- monosaccharide
- Any of a class of organic compounds which are the simplest forms of carbohydrates and the most basic structural subunits or monomers from which larger carbohydrate polymers such as disaccharides, oligosaccharides, and polysaccharides are composed. With few exceptions, all monosaccharides are variations on the empirical formula (CH
2O)
n, where n typically ranges from 3 (trioses) to 7 (heptoses).[3] Common examples include glucose, ribose, and deoxyribose. - monosomy
- The abnormal and frequently pathological presence of only one chromosome of a normal diploid pair. It is a type of aneuploidy.
- Morpholino
- A synthetic nucleic acid analogue connecting a short sequence of nucleobases into an artificial antisense oligomer, used in genetic engineering to knockdown gene expression by pairing with complementary sequences in naturally occurring RNA or DNA molecules, especially mRNA transcripts, thereby inhibiting interactions with other biomolecules such as proteins and ribosomes. Morpholino oligomers are not themselves translated, and neither they nor their hybrid duplexes with RNA are attacked by nucleases; also, unlike the negatively charged phosphates of normal nucleic acids, the synthetic backbones of Morpholinos are electrically neutral, making them less likely to interact non-selectively with a host cell's charged proteins. These properties make them useful and reliable tools for artificially generating mutant phenotypes in living cells.[13]
- mosaicism
- The presence of two or more populations of cells with different genotypes in an individual organism which has developed from a single fertilized egg. A mosaic organism can result from many kinds of genetic phenomena, including nondisjunction of chromosomes, endoreduplication, or mutations in individual stem cell lineages during the early development of the embryo. Mosaicism is similar to but distinct from chimerism.
- motif
- Any distinctive or recurring sequence of nucleotides in a nucleic acid or of amino acids in a peptide that is or is conjectured to be biologically significant, especially one that is reliably recognized by other biomolecules or which has a three-dimensional structure that permits unique or characteristic chemical interactions such as DNA binding.[13] In nucleic acids, motifs are often short (three to ten nucleotides in length), highly conserved sequences which act as recognition sites for DNA-binding proteins or RNAs involved in the regulation of gene expression.
- motor protein
- Any protein which converts chemical energy derived from the hydrolysis of nucleoside triphosphates such as ATP and GTP into mechanical work in order to effect its own locomotion, by propelling itself along a filament or through the cytoplasm.[4]
- mRNA
- See messenger RNA.
- mtDNA
- See mitochondrial DNA.
- multicellular
- Composed of more than one cell. The term is used especially to describe organisms or tissues consisting of many cells descendant from the same original parent cell which work together in an organized way, but may also describe groups of nominally single-celled organisms such as protists and bacteria which live symbiotically with each other in large colonies. Contrast unicellular.
- multimer
- An aggregate of two or more molecular entities, identical or non-identical, held together by non-covalent forces;[5] e.g. a protein complex.
- multinucleate
- (of a cell) Having more than one nucleus within a single cell; i.e. having multiple nuclei occupying the same cytoplasm.
- multiomics
- The integration of data from multiple "omics" technologies (e.g. data from the genome, epigenome, transcriptome, proteome, metabolome, etc.) in order to study complex biological relationships, discover novel associations between biological entities, pinpoint relevant biomarkers, or build elaborate models of physiology and disease.
- multiple cloning site (MCS)
- A locus or sequence within a plasmid vector which contains multiple unique restriction sites recognized by various restriction endonucleases, which makes it possible for scientists to target the insertion of a DNA fragment (often a gene cassette) specifically to that locus and in the desired orientation, by digesting the insert and the vector with the same endonuclease(s) and then ligating them together via compatible restriction ends, a technique known as restriction cloning.[5] Commercial plasmids designed for cloning commonly incorporate one or more multiple cloning sites.
- mutagen
- Any physical or chemical agent that changes the genetic material (usually DNA) of an organism and thereby increases the frequency of mutations above natural background levels.
- mutagenesis
- 1. The process by which the genetic information of an organism is changed, resulting in a mutation. Mutagenesis may occur spontaneously or as a result of exposure to a mutagen.
- 2. In molecular biology, any laboratory technique by which one or more genetic mutations are deliberately engineered in order to produce a mutant gene, regulatory element, gene product, or genetically modified organism so that the functions of a genetic locus, process, or product can be studied in detail.
- mutant
- An organism, gene product, or phenotypic trait resulting from a mutation, of a type that would not be observed naturally in wild-type specimens.
- mutation
- Any permanent change in the nucleotide sequence of a strand of DNA or RNA, or in the amino acid sequence of a peptide. Mutations play a role in both normal and abnormal biological processes; their natural occurrence is integral to the process of evolution. They can result from errors in replication, chemical damage, exposure to high-energy radiation, or manipulations by mobile genetic elements. Repair mechanisms have evolved in many organisms to correct them. By understanding the effect that a mutation has on phenotype, it is possible to establish the function of the gene or sequence in which it occurs.
- mutator gene
- Any mutant gene or sequence that increases the spontaneous mutation rate of one or more other genes or sequences. Mutators are often transposable elements, or may be mutant housekeeping genes such as those that encode helicases or proteins involved in proofreading.[13]
- mutein
- A mutant protein, i.e. a protein whose amino acid sequence differs from that of the normal because of a mutation.
- muton
- The smallest unit of a DNA molecule in which a physical or chemical change can result in a mutation (conventionally a single nucleotide).[13]



N
[edit]- n orientation
- One of two possible orientations by which a linear DNA fragment can be inserted into a vector, specifically the one in which the gene maps of both fragment and vector have the same orientation.[13] Contrast u orientation.
- NAD
- See nicotinamide-adenine dinucleotide.
- NADP
- See nicotinamide-adenine dinucleotide phosphate.
- nanoinjection
- A laboratory technique involving the use of a microscopic lance or nanopipette (typically about 100 nanometres in diameter) in the presence of an electric field in order to deliver DNA or RNA directly into a cell, often a zygote or early embryo, via an electrophoretic mechanism. While submerged in a pH-buffered solution, a positive electric charge is applied to the lance, attracting negatively charged nucleic acids to its surface; the lance then penetrates the cell membrane and the electric field is reversed, applying a negative charge which repels the accumulated nucleic acids away from the lance and thus into the cell. Compare microinjection.
- nascent
- In the process of being synthesized; incomplete; not yet fully processed or mature. The term is commonly used to describe strands of DNA or RNA which are actively undergoing synthesis during replication or transcription, or sometimes a complete, fully transcribed RNA molecule before any post-transcriptional modifications have been made (e.g. polyadenylation or RNA editing), or a peptide chain actively undergoing translation by a ribosome.[13]
- ncAA
- See non-canonical amino acid.
- ncDNA
- See non-coding DNA.
- ncRNA
- See non-coding RNA.
- negative (-) sense strand
- See template strand.
- negative control
- The inhibition or deactivation of some biological process caused by the presence of a specific molecular entity (e.g. a repressor), in the absence of which the process is not inhibited and thus can proceed normally.[5] In gene regulation, for example, a repressor may bind to an operator upstream from a coding sequence and prevent access by transcription factors and/or RNA polymerase, thereby blocking the gene's transcription. This is contrasted with positive control, in which the presence of an inducer is necessary to switch on transcription.[9]
- negative supercoiling
- The supercoiling of a double-stranded DNA molecule in the direction opposite to the turn of the double helix itself (e.g. a left-handed coiling of a helix with a right-handed turn).[9] Contrast positive supercoiling.
- next-generation sequencing (NGS)
- See massively parallel sequencing.
- nick
- A break or discontinuity in the phosphate backbone of one strand of a double-stranded DNA molecule, i.e. where a phosphodiester bond is hydrolyzed but no nucleotides are removed; such a molecule is said to be nicked. A nick is a single-strand break, where despite the break the DNA molecule is not ultimately broken into multiple fragments, which contrasts with a cut, where both strands are broken. Nicks may be caused by DNA damage or by dedicated nucleases known as nicking enzymes, which nick DNA at random or specific sites. Nicks are frequently placed by the cell as markers identifying target sites for enzyme activity, including in DNA replication, transcription, and mismatch repair, and also to release torsional stress from overwound DNA molecules, making them important in manipulating DNA topology.[9]
- nick translation
- nickase
- Another name for a nicking enzyme, especially one that has been artificially engineered to create single-stranded breaks (i.e. nicks) by altering the cleavage activity of an endonuclease that normally creates double-stranded breaks, e.g. Cas9 nickase (nCas9).[15]
- nicking enzyme
- Any of a class of endonuclease enzymes capable of generating a single-stranded break in a double-stranded DNA molecule, i.e. a nick, either at random or at a specific recognition sequence, by breaking a phosphodiester bond linking adjacent nucleotides.
- nicotinamide adenine dinucleotide (NAD)
- nicotinamide adenine dinucleotide phosphate (NADP+, NADP)
- nitrogenous base
- Any organic compound containing a nitrogen atom that has the chemical properties of a base. Five particular nitrogenous bases – adenine (A), guanine (G), cytosine (C), thymine (T), and uracil (U) – are especially relevant to biology because they are components of nucleotides, which are the primary monomers that make up nucleic acids.
- non-canonical amino acid (ncAA)
- Any amino acid, natural or artificial, that is not one of the 20 or 21 proteinogenic amino acids encoded by the standard genetic code. There are hundreds of such amino acids, many of which have biological functions and are specified by alternative codes or incorporated into proteins accidentally by errors in translation. Many of the best known naturally occurring ncAAs occur as intermediates in the metabolic pathways leading to the standard amino acids, while others have been made synthetically in the laboratory.[16]
- non-coding DNA (ncDNA)
- Any segment of DNA that does not encode a sequence that may ultimately be transcribed and translated into a protein. In most organisms, only a small fraction of the genome consists of protein-coding DNA, though the proportion varies greatly between species. Some non-coding DNA may still be transcribed into functional non-coding RNA (as with transfer RNAs) or may serve important developmental or regulatory purposes; other regions (as with so-called "junk DNA") appear to have no known biological function.
- non-coding RNA (ncRNA)
- Any molecule of RNA that is not ultimately translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often referred to as an "RNA gene". Numerous types of non-coding RNAs essential to normal genome function are produced constitutively, including transfer RNA (tRNA), ribosomal RNA (rRNA), microRNA (miRNA), and small interfering RNA (siRNA); other non-coding RNAs (sometimes described as "junk RNA") have no known function and are likely the product of spurious transcription.
- non-coding strand
- See template strand.
- nondisjunction
- The failure of homologous chromosomes or sister chromatids to segregate properly during cell division. Nondisjunction results in daughter cells that are aneuploid, containing abnormal numbers of one or more specific chromosomes. It may be caused by a variety of factors.
- non-homologous end joining (NHEJ)
- nonrepetitive sequence
- Broadly, any nucleotide sequence or region of a genome that does not contain repeated sequences, or in which repeats do not comprise a majority; or any segment of DNA exhibiting the reassociation kinetics expected of a unique sequence.[13]
- nonsense mutation
- A type of point mutation which results in a premature stop codon in the transcribed mRNA sequence, thereby causing the premature termination of translation, which results in a truncated, incomplete, and often non-functional protein.
- nonsense suppressor
- A factor which can inhibit the effects of a nonsense mutation (i.e. a premature stop codon) by any mechanism, usually either a mutated transfer RNA which can bind the mutated stop codon or some kind of ribosomal mutation.[17]
- nonsynonymous mutation
- A type of mutation in which the substitution of one nucleotide base for another results, after transcription and translation, in an amino acid sequence that is different from that produced by the original unmutated gene. Because nonsynonymous mutations always result in a biological change in the organism, they are often subject to strong selection pressure. Contrast synonymous mutation.
- non-transcribed spacer (NTS)
- See spacer.
- northern blotting
- A blotting method in molecular biology used to detect RNA in a sample. Compare Southern blotting, western blotting, and eastern blotting.
- nRNA
- See nuclear RNA.
- N-terminus
- The end of a linear chain of amino acids (i.e. a peptide) that is terminated by the free amine group (–NH
2) of the first amino acid added to the chain during translation. This amino acid is said to be N-terminal. By convention, sequences, domains, active sites, or any other structure positioned nearer to the N-terminus of the polypeptide or the folded protein it forms relative to others are described as upstream. Contrast C-terminus. - nuclear DNA
- Any DNA molecule contained within the nucleus of a eukaryotic cell, most prominently the DNA in chromosomes. It is sometimes used interchangeably with genomic DNA.
- nuclear envelope
- A sub-cellular barrier consisting of two concentric lipid bilayer membranes that surrounds the nucleus in eukaryotic cells. The nuclear envelope is sometimes simply called the "nuclear membrane", though the structure is actually composed of two distinct membranes, an inner membrane and an outer membrane.
- nuclear equivalence
- The principle that the nuclei of essentially all differentiated cells of a mature multicellular organism are genetically identical to each other and to the nucleus of the zygote from which they descended; i.e. they all contain the same genetic information on the same chromosomes, having been replicated from the original zygotic set with extremely high fidelity. Even though all adult somatic cells have the same set of genes, cells can nonetheless differentiate into distinct cell types by expressing different subsets of these genes. Though this principle generally holds true, the reality is slightly more complex, as mutations such as insertions, deletions, duplications, and translocations as well as chimerism, mosaicism, and various types of genetic recombination can all cause different somatic lineages within the same organism to be genetically non-identical.
- nuclear export signal (NES)
- nuclear lamina
- A fibrous network of proteins lining the inner, nucleoplasmic surface of the nuclear envelope, composed of filaments similar to those that make up the cytoskeleton. It may function as a scaffold for the various contents of the nucleus including nuclear proteins and chromosomes.[3]
- nuclear localization signal (NLS)
- An amino acid sequence within a protein which serves as a molecular signal marking the protein for transport into the nucleus, typically consisting of one or more short motifs containing positively charged amino acid residues exposed on the mature protein's surface (especially lysines and arginines). Though all proteins are translated in the cytoplasm, many whose primary biological activities occur inside the nucleus (e.g. transcription factors) require nuclear localization signals identifiable by molecular chaperones in order to cross the nuclear envelope. Contrast nuclear export signal.
- nuclear matrix
- A mesh-like latticework of protein polymers and microfilaments suspended in the nucleoplasm in the nuclei of eukaryotic cells, akin to the cytoskeleton in the cytoplasm. The nuclear matrix functions as a scaffold and an anchor for large DNA molecules such as chromosomes and for the macromolecular complexes that perform essential nuclear activities such as transcription and DNA replication.[3]
- nuclear membrane
- See nuclear envelope.
- nuclear pore
- A complex of membrane proteins that creates an opening in the nuclear envelope through which certain molecules and ions are permitted to pass and thereby enter or exit the nucleus (analogous to the channel proteins in the cell membrane). The nuclear envelope typically has thousands of pores to selectively regulate the exchange of specific materials between the nucleoplasm and the cytoplasm, including messenger RNAs, which are transcribed in the nucleus but must be translated in the cytoplasm, as well as nuclear proteins, which are synthesized in the cytoplasm but must return to the nucleus to serve their functions.[4][3]
- nuclear protein
- Any protein that is naturally found in or localizes to the cell's nucleus (as opposed to the cytoplasm or elsewhere).
- nuclear RNA (nRNA)
- Any RNA molecule located within a cell's nucleus, whether associated with chromosomes or existing freely in the nucleoplasm, including small nuclear RNA (snRNA), enhancer RNA (eRNA), and all newly transcribed immature RNAs, coding or non-coding, prior to their export to the cytosol (hnRNA).
- nuclear transfer
- nuclear transport
- The mechanisms by which molecules cross the nuclear envelope surrounding a cell's nucleus. Though small molecules and ions can cross the membrane freely, the entry and exit of larger molecules is tightly regulated by nuclear pores, so that most macromolecules such as RNAs and proteins require association with transport factors in order to be chaperoned across.
- nuclease
- Any of a class of enzymes capable of cleaving phosphodiester bonds connecting adjacent nucleotides in a nucleic acid molecule (the opposite of a ligase). Nucleases may nick one strand or cut both strands of a duplex molecule, and may cleave randomly or at specific recognition sequences. They are ubiquitous and imperative for normal cellular function, and are also widely employed in laboratory techniques.
- nucleic acid
- A long, polymeric macromolecule made up of smaller monomers called nucleotides which are chemically linked to one another in a chain. Two specific types of nucleic acid, DNA and RNA, are common to all living organisms, serving to encode the genetic information governing the construction, development, and ordinary processes of all biological systems. This information, contained within the order or sequence of the nucleotides, is translated into proteins, which direct all of the chemical reactions necessary for life.
- nucleic acid sequence
- The precise order of consecutively linked nucleotides in a nucleic acid molecule such as DNA or RNA. Long sequences of nucleotides are the principal means by which biological systems store genetic information, and therefore the accurate replication, transcription, and translation of such sequences is of the utmost importance, lest the information be lost or corrupted. Nucleic acid sequences may be equivalently referred to as sequences of nucleotides, nitrogenous bases, nucleobases, or, in duplex molecules, base pairs, and they correspond directly to sequences of codons and amino acids.
- nucleobase
- Any of the five primary or canonical nitrogenous bases – adenine (A), guanine (G), cytosine (C), thymine (T), and uracil (U) – that form nucleosides and nucleotides, the latter of which are the fundamental building blocks of nucleic acids. The ability of these bases to form base pairs via hydrogen bonding, as well as their flat, compact three-dimensional profiles, allows them to "stack" one upon another and leads directly to the long-chain structures of DNA and RNA. When writing sequences in shorthand notation, the letter N is often used to represent a nucleotide containing a generic or unidentified nucleobase.
- nucleoid
- An irregularly shaped region which contains most or all of the genetic material in prokaryotic cells such as bacteria, but is not enclosed by a nuclear membrane as in eukaryotes.
- nucleolin
- The primary protein of which the eukaryotic nucleolus is composed, thought to play important roles in chromatin decondensation, transcription of ribosomal RNA, and ribosome assembly.
- nucleolonema
- The central region of the nucleolus, composed of dense, convoluted fibrillar material.[2]
- nucleolus
- An organelle within the nucleus of eukaryotic cells which is composed of proteins, DNA, and RNA and serves as the site of ribosome synthesis.
- nucleoplasm
- All of the material enclosed within the nucleus of a cell by the nuclear envelope, analogous to the cytoplasm enclosed by the main cell membrane. Like the cytoplasm, the nucleoplasm is composed of a gel-like substance (the nucleosol) in which various organelles, nuclear proteins, and other biomolecules are suspended, including nuclear DNA in the form of chromosomes, the nucleolus, nuclear bodies, and free nucleotides.
- nucleoprotein
- Any protein that is chemically bonded to or conjugated with a nucleic acid. Examples include ribosomes, nucleosomes, and many enzymes.
- nucleosidase
- Any of a class of enzymes which catalyze the decomposition of nucleosides into their component nitrogenous bases and pentose sugars.[13]
- nucleoside
- An organic molecule composed of a nitrogenous base bonded to a five-carbon sugar (either ribose or deoxyribose). A nucleotide additionally includes one or more phosphate groups.
- nucleosol
- The soluble, liquid portion of the nucleoplasm (analogous to the cytosol of the cytoplasm).
- nucleosome
- The basic structural subunit of chromatin used in packaging nuclear DNA such as chromosomes, consisting of a core particle of eight histone proteins around which double-stranded DNA is wrapped in a manner akin to thread wound around a spool. The technical definition of a nucleosome includes a segment of DNA about 146 base pairs in length which makes 1.67 left-handed turns as it coils around the histone core, as well as a stretch of linker DNA (generally 38–80 bp) connecting it to an adjacent core particle, though the term is often used to refer to the core particle alone. Long series of nucleosomes are further condensed by association with histone H1 into higher-order structures such as 30-nm fibers and ultimately supercoiled chromatids. Because the histone–DNA interaction limits access to the DNA molecule by other proteins and RNAs, the precise positioning of nucleosomes along the DNA sequence plays a fundamental role in controlling whether or not genes are transcribed and expressed, and hence mechanisms for moving and ejecting nucleosomes have evolved as a means of regulating the expression of particular loci.
- nucleosome-depleted region (NDR)
- A region of a genome or chromosome in which long segments of DNA are bound by few or no nucleosomes, and thus exposed to manipulation by other proteins and molecules, especially implying that the region is transcriptionally active.
- nucleotidase
- Any of a class of phosphoric monoester hydrolase enzymes which catalyze the hydrolysis of a nucleotide into a nucleoside and orthophosphate. Such enzymes may or may not distinguish ribonucleotides from deoxyribonucleotides, i.e. their function may not be sugar-specific.[5]
- nucleotide
- An organic molecule that serves as the fundamental monomer or subunit of nucleic acid polymers, including RNA and DNA. Each nucleotide is composed of three connected functional groups: a nitrogenous base, a five-carbon sugar (either ribose or deoxyribose), and a single phosphate group. Though technically distinct, the term "nucleotide" is often used interchangeably with nitrogenous base, nucleobase, and base pair when referring to the sequences that make up nucleic acids. Compare nucleoside.
- nucleotide sequence
- See nucleic acid sequence.
- nucleus
- A large spherical or lobular organelle surrounded by a dedicated membrane which functions as the main storage compartment for the genetic material of eukaryotic cells, including the DNA comprising chromosomes, as well as the site of RNA synthesis during transcription. The vast majority of eukaryotic cells have a single nucleus, though some cells may have more than one nucleus, either temporarily or permanently, and in some organisms there exist certain cell types (e.g. mammalian erythrocytes) which lose their nuclei upon reaching maturity, effectively becoming anucleate. The nucleus is one of the defining features of eukaryotes; the cells of prokaryotes such as bacteria lack nuclei entirely.[2]

O
[edit]- occluding junction
- See tight junction.
- ochre
- One of three stop codons used in the standard genetic code; in RNA, it is specified by the nucleotide triplet UAA. The other two stop codons are named amber and opal.
- Okazaki fragments
- Short sequences of nucleotides which are synthesized discontinuously by DNA polymerase and later linked together by DNA ligase to create the lagging strand during DNA replication. Okazaki fragments are the consequence of the unidirectionality of DNA polymerase, which only works in the 5' to 3' direction.
- oligo dT
- A short, single-stranded DNA oligonucleotide consisting of a sequence of repeating deoxythymidine (dT) nucleotides. Oligo dTs are commonly synthesized de novo to be used as primers for in vitro reverse transcription reactions during rtPCR techniques, where short chains of 12 to 18 thymine bases readily complement the poly(A) tails of mature messenger RNAs, allowing the selective amplification and preparation of a cDNA library from a pool of coding transcripts.[5]
- oligogene
- oligomer
- Any polymeric molecule consisting of a relatively short series of connected monomers or subunits; e.g. an oligonucleotide is a short series of nucleotides.
- oligonucleotide
- A relatively short chain of nucleic acid residues. In the laboratory, oligonucleotides are commonly used as primers or hybridization probes to detect the presence of larger mRNA molecules or assembled into two-dimensional microarrays for high-throughput sequencing analysis.
- oligosaccharide
- A polymeric carbohydrate molecule consisting of a relatively short chain of connected monosaccharides. Oligosaccharides have important functions in processes such as cell signaling and cell adhesion. Longer chains are called polysaccharides.
- omics
- A suffix used to describe any of the diverse fields of study that conduct rigorous, systematic analyses of any of the "omes", e.g. the genome, transcriptome, proteome, metabolome, etc.,[5] each of which represents the totality of a specific class of biological content that has been or could hypothetically be isolated from an individual cell, population of cells, organism, species, or some other particular context. Thus genomics is the field of study which analyzes the totality of genes in a genome, proteomics studies the complete set of all of the proteins in a proteome, etc. The term may also be used to refer to all of these fields collectively.
- oncogene
- A gene that has the potential to cause cancer. In tumor cells, such genes are often mutated and/or expressed at abnormally high levels.
- one gene–one polypeptide
- The hypothesis that there exists a large class of genes in which each particular gene directs the synthesis of one particular polypeptide or protein.[13] Historically it was thought that all genes and proteins might follow this rule by definition, but it is now known that many proteins are composites of different polypeptides and therefore the product of multiple genes, and also that some genes do not encode polypeptides at all but instead produce non-coding RNAs, which are never translated.
- opal
- One of three stop codons used in the standard genetic code; in RNA, it is specified by the nucleotide triplet UGA. The other two stop codons are named amber and ochre.
- open chromatin
- See euchromatin.
- open reading frame (ORF)
- The part of a reading frame that has the ability to be translated from DNA or RNA into protein; any continuous stretch of codons that contains a start codon and a stop codon.
- operator
- A regulatory sequence within an operon, typically located between the promoter sequence and the structural genes of the operon, to which an uninhibited repressor protein can bind, thereby physically obstructing RNA polymerase from initiating the transcription of adjacent cistrons.[18]
- operon
- A functional unit of gene expression consisting of a cluster of neighboring structural genes which are collectively under the control of a single promoter, along with one or more adjacent regulatory sequences such as operators. The set of genes is transcribed together, resulting in a single polycistronic messenger RNA molecule encoding multiple distinct polypeptides which may then be translated together or undergo splicing to create multiple mRNAs which are translated independently; the result is that the genes contained in the operon are either expressed together or not at all. Regulatory proteins, including repressors and activators, usually bind specifically to the regulatory sequences of a given operon; by some definitions, the genes that code for these regulatory proteins are also considered part of the operon.
- opsonin
- Any substance, especially certain blood-serum proteins such as immunoglobulins, that in binding to the surface of foreign cells or particulate matter increases the susceptibility of the foreign material to phagocytosis by phagocytes.[3] Opsonins work by linking foreign particles to specific receptors on the surface of phagocytic cells in a process known as opsonization.[5]
- organelle
- A spatially distinct compartment or subunit within a cell which has a specialized function. Organelles occur in both prokaryotic and eukaryotic cells. In the latter they are often separated from the cytoplasm by being enclosed with their own membrane bilayer (whence the term membrane-bound organelles), though organelles may also be functionally specific areas or structures without a surrounding membrane; some cellular structures which exist partially or entirely outside of the cell membrane, such as cilia and flagella, are also referred to as organelles. There are numerous types of organelles with a wide variety of functions, including the various compartments of the endomembrane system (e.g. the nuclear envelope, endoplasmic reticulum, and Golgi apparatus), mitochondria, chloroplasts, lysosomes, endosomes, and vacuoles, among others. Many organelles are unique to particular cell types or species.
- origin of replication (ORI)
- A particular location within a DNA molecule at which DNA replication is initiated. Origins are usually defined by the presence of a particular replicator sequence or by specific chromatin patterns.
- osmosis
- osmotic shock
- Physiological dysfunction caused by a sudden change in the concentration of dissolved solutes in the extracellular environment surrounding a cell, which provokes the rapid movement of water across the cell membrane by osmosis, either into or out of the cell. In a severely hypertonic environment, where extracellular solute concentrations are extremely high, osmotic pressure may force large quantities of water to move out of the cell (plasmolysis), leading to its desiccation; this may also have the effect of inhibiting transport of solutes into the cell, thus denying it the substrates necessary to sustain normal cellular activities. In a severely hypotonic environment, where extracellular solute concentrations are much lower than intracellular concentrations, water is forced to move into the cell (turgescence), causing it to swell in size and potentially burst, or triggering apoptosis.
- outron
- A sequence near the 5'-end of a primary mRNA transcript that is removed by a special form of splicing during post-transcriptional processing. Outrons are located entirely outside of the transcript's coding sequences, unlike introns.
- overexpression
- An abnormally high level of gene expression which results in an excessive number of copies of one or more gene products. Overexpression produces a pronounced gene-related phenotype.[19][20]
- oxidative phosphorylation
- The process by which cells use chemical energy obtained by the oxidation of nutrients to power the production of adenosine triphosphate (ATP). Oxidative phosphorylation couples two related processes: in the electron transport chain, a series of enzyme-catalyzed redox reactions transfers electrons from energetic donors such as NADH and FADH through various intermediates and ultimately to a terminal electron acceptor such as molecular oxygen (O
2); the energy liberated by these reactions is simultaneously used in chemiosmosis to move protons (H+
) across a membrane and against their concentration gradient, generating an electrochemical potential which powers ATP synthase, an enzyme that catalyzes the phosphorylation of ADP into ATP. In eukaryotes, both of these processes are carried out by proteins embedded in the membranes of mitochondria and chloroplasts; in prokaryotes, they occur in the cell membrane. - oxidative stress
- oxygen cascade
- The flow of oxygen from environmental sources (e.g. the air in the atmosphere) to the mitochondria of a cell, where oxygen atoms participate in biochemical reactions that result in the oxidation of energy-rich substrates such as carbohydrates in a process known as aerobic respiration.
P
[edit]- p53
- A class of regulatory proteins encoded by the TP53 gene in vertebrates which bind DNA and regulate gene expression in order to protect the genome from mutation and block progression through the cell cycle if DNA damage does occur.[4] It is mutated in more than 50% of human cancers, indicating it plays a crucial role in preventing cancer formation.
- pachynema
- In meiosis, the third of five substages of prophase I, following zygonema and preceding diplonema. During pachynema, the synaptonemal complex facilitates crossing over between the synapsed homologous chromosomes, and the centrosomes begin to move apart from each other.[13]
- palindromic sequence
- A nucleic acid sequence of a double-stranded DNA or RNA molecule in which the unidirectional sequence (e.g. 5' to 3') of nucleobases on one strand is identical to the sequence in the same direction (e.g. 5' to 3') on the complementary strand. In other words, a sequence is said to be palindromic if it is equal to its own reverse complement. Palindromic motifs are common recognition sites for restriction enzymes.
- paracellular transport
- The transfer of substances across an epithelium by passing through the extracellular space between cells, in contrast to transcellular transport, where substances travel through cells by crossing the intracellular cytoplasm.
- paracrine
- Describing or relating to a class of agonist signaling molecules produced and secreted by regulatory cells into the extracellular environment and then transported by passive diffusion to target cells other than those which produced them. The term may refer to the molecules themselves, sometimes called paramones, to the cells that produce them, or to signaling pathways which rely on them.[5] Compare autocrine, endocrine, and juxtacrine.
- paratope
- An idiotope, i.e. the specific site or region within an antibody that recognizes and binds to a particular antigen or epitope.[21] The uniqueness of a paratope allows it to bind to only one epitope with very high affinity. At the end of each arm of the Y-shaped antibody is an identical paratope, and each paratope comprises a total of six complementarity-determining regions (three from each of the light and heavy chains) which protrude from a series of antiparallel beta sheets in the antibody's higher structure.[22] The term is also sometimes used to refer to the specific site on a ligand molecule which defines the ligand's specificity for other molecules such as cell-surface receptors.[3]
- parent cell
- The original or ancestral cell from which a given set of descendant cells, known as daughter cells, have divided by mitosis or meiosis.
- passenger
- A DNA fragment of interest designed to be spliced into a 'vehicle' such as a plasmid vector and then cloned.[13]
- passive transport
- The movement of a solute across a membrane by traveling down an electrochemical or concentration gradient, using only the energy stored in the gradient and not any energy from external sources.[3] Contrast active transport.
- Pasteur effect
- A phenomenon observed in facultatively anaerobic cells, including animal tissues and many microorganisms such as yeast, whereby the presence of oxygen in the environment inhibits the cell's use of ethanol fermentation pathways to generate energy, and drives the cell to instead make use of the available oxygen in aerobic respiration;[23] or more generally the observed decrease in the rate of glycolysis or of lactate production in cells exposed to oxygenated air.[5]
- PCR
- See polymerase chain reaction.
- PCR product
- See amplicon.
- pentose
- Any monosaccharide containing five carbon atoms. The compounds ribose and deoxyribose are both pentose sugars, which, in the form of cyclic five-membered rings, serve as the central structural components of the ribonucleotides and deoxyribonucleotides that make up RNA and DNA, respectively.
- peptidase
- See protease.
- peptide
- A short chain of amino acid monomers linked by covalent peptide bonds. Peptides are the fundamental building blocks of longer polypeptide chains and hence of proteins.
- peptide bond
- A covalent chemical bond between the carboxyl group of one amino acid and the amino group of an adjacent amino acid, formed by a dehydration reaction catalyzed by peptidyl transferase, an enzyme within the ribosome, during translation. A linear chain of amino acids linked by peptide bonds may be called a peptide or polypeptide.
- peptidoglycan
- A glycoconjugate complex of interwoven peptides and polysaccharides that is a primary constituent of the cell wall in all bacteria and archaea, consisting of strands of glycosaminoglycans crosslinked by short oligopeptides (usually 4–10 residues[3]) to form a rigid lattice of indefinite size.[5] The proportion of the cell wall that is peptidoglycan varies widely by strain and is often used to aid strain identification: the higher peptidoglycan content of the cell walls of Gram-positive bacteria causes them to stain a darker color than Gram-negative bacteria.
- pericentriolar material (PCM)
- perinuclear space
- The space between the inner and outer membranes of the nuclear envelope.
- peripheral membrane protein
- Any of a class of membrane proteins which attach only temporarily to the cell membrane, either by penetrating the lipid bilayer or by attaching to other proteins which are permanently embedded within the membrane.[24] The ability to reversibly interact with membranes makes peripheral membrane proteins important in many different roles, commonly as regulatory subunits of channel proteins and cell surface receptors. Their domains often undergo rearrangement, dissociation, or conformational changes when they interact with the membrane, resulting in the activation of their biological activity.[25] In protein purification, peripheral membrane proteins are typically more water-soluble and much easier to isolate from the membrane than integral membrane proteins.
- periplasmic space
- In a bacterial cell, the space between the cell membrane and the cell wall.[5]
- peroxisome
- A small membrane-bound organelle found in many eukaryotic cells which specializes in carrying out oxidative reactions with various enzyme peroxidases and catalase, generally to mitigate damage from reactive oxygen species but also as a participant in various metabolic pathways such as beta-oxidation of fatty acids.[26]
- persistence
- 1. The tendency of a moving cell to continue moving in the same direction as previously; that is, even in isotropic environments, there inevitably still exists an inherent bias by which, from instant to instant, cells are more likely not to change direction than to change direction. Averaged over long periods of time, however, this bias is less obvious and cell movements are better described as a random walk.[3]
- 2. The ability of some viruses to remain present and viable in cells, organisms, or populations for very long periods of time by any of a variety of strategies, including retroviral integration and immune suppression, often in a latent form which replicates very slowly or not at all.[3]
- Petri dish
- A shallow, transparent plastic or glass dish, usually circular and covered with a lid, which is widely used in biology laboratories to hold solid or liquid growth media for the purpose of culturing cells. They are particularly useful for adherent cultures, where they provide a flat, sterile surface conducive to colony formation from which scientists can easily isolate and identify individual colonies.
- phagemid
- A bacteriophage with a genome encoding a mobile plasmid that can be excised by co-infection of the host cell with a helper phage. Phagemids are useful as vectors for library production.[3]
- phagocyte
- Any cell capable of phagocytosis, especially any of various cell types of the immune system which engulf and ingest harmful foreign molecules, bacteria, and dead or dying cells, including neutrophils and macrophages.[3]
- phagocytosis
- The process by which foreign cells, molecules, and small particulate matter are engulfed and ingested via endocytosis by specialized cells known as phagocytes (a class which includes macrophages and neutrophils).[4]
- phagosome
- A large, intracellular, membrane-bound vesicle formed as a result of phagocytosis and containing whatever previously extracellular material was engulfed during that process.[4]
- pharmacogenomics
- The study of the role played by the genome in the body's response to pharmaceutical drugs, combining the fields of pharmacology and genomics.
- phenome
- The complete set of phenotypes that are or can be expressed by a genome, cell, tissue, organism, or species; the sum of all of its manifest chemical, morphological, and behavioral characteristics or traits.
- phenomic lag
- A delay in the phenotypic expression of a genetic mutation owing to the time required for the manifestation of changes in the affected biochemical pathways.[9]
- phenotype
- The composite of the observable morphological, physiological, and behavioral traits of an organism that result from the expression of the organism's genotype as well as the influence of environmental factors and the interactions between the two.
- phenotypic switching
- A type of phenotypic plasticity in which a cell rapidly undergoes major changes to its morphology and/or function, usually via epigenetic modifications, allowing it to quickly switch back and forth between disparate phenotypes in response to changes in the local microenvironment.
- phosphatase
- Any of a class of enzymes that catalyze the hydrolytic cleavage of a phosphoric acid monoester into a phosphate ion and an alcohol, e.g. the removal of a phosphate group from a nucleotide via the breaking of the ester bond connecting the phosphate to a ribose or deoxyribose sugar or to another phosphate, a process termed dephosphorylation. The opposite process is performed by kinases.
- phosphate
- Any chemical species or functional group derived from phosphoric acid (H
3PO
4) by the removal of one or more protons (H+
); the completely ionized form, [PO
4]3−
, consists of a single, central phosphorus atom covalently bonded to four oxygen atoms via three single bonds and one double bond. Phosphates are abundant and ubiquitous in biological systems, where they occur either as free anions in solution, known as inorganic phosphates and symbolized Pi, or bonded to organic molecules via ester bonds. The huge diversity of organophosphate compounds includes all nucleotides, whose phosphate groups are linked by phosphodiester bonds to form the structural backbones of long nucleotide chains such as DNA and RNA, and the high-energy diphosphate and triphosphate substituents of individual nucleotides such as ADP and ATP serve as essential energy carriers in all cells. Phospholipids are major components of most membranes. Enzymes known as kinases and phosphatases catalyze the addition and removal of phosphate groups to and from these and other biomolecules. - phosphate backbone
- The linear chain of alternating phosphate and sugar compounds that results from the linking of consecutive nucleotides in the same strand of a nucleic acid molecule, and which serves as the structural framework of the nucleic acid. Each individual strand is held together by a repeating series of phosphodiester bonds connecting each phosphate group to the ribose or deoxyribose sugars of two adjacent nucleotides. These bonds are created by ligases and broken by nucleases.
- phosphodiester bond
- A pair of ester bonds linking a phosphate molecule with the two pentose rings of consecutive nucleosides on the same strand of a nucleic acid. Each phosphate forms a covalent bond with the 3' carbon of one pentose and the 5' carbon of the adjacent pentose; the repeated series of such bonds that holds together the long chain of nucleotides comprising DNA and RNA molecules is known as the phosphate or phosphodiester backbone.
- phospholipid
- Any of a subclass of lipids consisting of a central alcohol (usually glycerol) covalently bonded to three functional groups: a negatively charged phosphate group, and two long fatty acid chains. This arrangement results in a highly amphipathic molecule which in aqueous solutions tends to aggregate with similar molecules in a lamellar or micellar conformation with the hydrophilic phosphate "heads" oriented outward, exposing them to the solution, and the hydrophobic fatty acid "tails" oriented inward, minimizing their interactions with water and other polar compounds. Phospholipids are the major structural membrane lipid in almost all biological membranes except the membranes of some plant cells and chloroplasts, where glycolipids dominate instead.[3]
- phospholipid bilayer
- See lipid bilayer.
- phosphorylation
- The attachment of a phosphate ion, PO3−
4, to another molecule or ion or to a protein by covalent bonding. Phosphorylation and the inverse reaction, dephosphorylation, are essential steps in numerous biochemical pathways, including in the production of adenosine triphosphate (ATP) (as in oxidative phosphorylation); in the metabolism of glucose and the synthesis of glycogen; and in the post-translational modification of amino acid residues in many proteins. Enzymes which catalyze phosphorylation reactions are known as kinases; those that catalyze dephosphorylation are known as phosphatases. - pinocytosis
- A form of endocytosis in which liquid and suspended solids from the extracellular environment are captured in inward invaginations of the cell membrane which then "bud off" into enclosed vesicles in the cytoplasm. The contents of these vesicles are then passed to organelles such as endosomes by fusion of the vesicular and organellar membranes. Pinocytosis is the predominant form of endocytosis occurring in most cells, such that the term is often used interchangeably with endocytosis as a whole.[5]
- piRNA
- See Piwi-interacting RNA.
- pitch
- The number of base pairs contained within a single complete turn of the DNA double helix,[13] used as a measure of the "tightness" or density of the helix's spiral.
- Piwi-interacting RNA (piRNA)
- plasma membrane
- See cell membrane.
- plasmid
- Any small DNA molecule that is physically separated from the larger body of chromosomal DNA and can replicate independently. Plasmids are typically small (less than 100 kbp), circular, double-stranded DNA molecules in prokaryotes such as bacteria, though they are also sometimes present in archaea and eukaryotes.
- plasmid partitioning
- The process by which plasmids which have been replicated inside a parent cell are distributed equally between daughter cells during cell division.[3]
- plasmid-mediated resistance
- The development of resistance to toxins or antibiotics which is enabled by the horizontal transfer of resistance genes encoded within small, independently replicating DNA molecules known as plasmids. This process occurs naturally via mechanisms such as bacterial conjugation, but is also a common aspect of genetic engineering methods such as molecular cloning.
- plasmolysis
- The temporary shrinkage of the protoplasm of a plant or bacterial cell away from the cell wall, caused by loss of water from the cell.[5]
- plastid
- Any of a class of membrane-bound organelles found in the cells of some eukaryotes such as plants and algae which are hypothesized to have evolved from endosymbiotic cyanobacteria; examples include chloroplasts, chromoplasts, and leucoplasts. Plastids retain their own circular chromosomes which replicate independently of the host cell's genome. Many contain photosynthetic pigments which allow them to perform photosynthesis, while others have been retained for their ability to synthesize unique chemical compounds.
- pleomorphism
- 1. Variability in the size, shape, or staining of cells and/or their nuclei, particularly as observed in histology and cytopathology, where morphological variation is frequently an indicator of a cellular abnormality such as disease or tumor formation.
- 2. In microbiology, the ability of some microorganisms such as certain bacteria and viruses to alter their morphology, metabolism, or mode of reproduction in response to changes in their environment.
- plithotaxis
- The tendency of cells within a monolayer to migrate in the direction of the local highest tension or maximal principal stress, exerting minimal shear stress on neighboring cells and thereby propagating the tension across many intercellular junctions and causing the cells to exhibit a sort of collective migration.[27]
- ploidy
- The number of complete sets of chromosomes in a cell, and hence the number of possible alleles present within the cell at any given autosomal locus.
- pluripotency
- plus-strand
- See coding strand.
- point mutation
- A mutation by which a single nucleotide base is changed, inserted, or deleted from a sequence of DNA or RNA.
- poly(A) tail
- A post-transcriptional modification consisting of a chain of repeated adenosine residues, 40–250 nucleotides in length, attached to the 3' end of nearly all mature eukaryotic messenger RNA transcripts (those of histones being a notable exception).[5]
- polyadenylation
- The addition of a series of multiple adenosine ribonucleotides, known as a poly(A) tail, to the 3'-end of a primary RNA transcript, typically a messenger RNA. Primary transcripts are first cleaved 10–30 nucleotides downstream of a highly conserved AAUAAA sequence, then the poly(A) tail is generated from the chaining of multiple ATP molecules through the action of polynucleotide adenylyltransferase.[5] A class of post-transcriptional modification, polyadenylation serves different purposes in different cell types and organisms. In eukaryotes, the addition of a poly(A) tail is an important step in the processing of a raw transcript into a mature mRNA, ready for export to the cytoplasm where translation occurs; in many bacteria, polyadenylation has the opposite function, instead promoting the RNA's degradation.
- polyclonal
- Describing cells, proteins, or molecules descended or derived from more than one clone (i.e. from more than one genome or genetic lineage) or made in response to more than one unique stimulus. Antibodies are often described as polyclonal if they have been produced or raised against multiple distinct antigens or multiple variants of the same antigen, such that they can recognize more than one unique epitope.[2] Contrast monoclonal.
- polylinker
- See multiple cloning site.
- polymer
- A macromolecule composed of multiple repeating subunits or monomers; a chain or aggregation of many individual molecules of the same compound or class of compound.[2] The formation of polymers is known as polymerization and generally only occurs when nucleation sites are present and the concentration of monomers is sufficiently high.[3] Many of the major classes of biomolecules are polymers, including nucleic acids and polypeptides.
- polymerase
- Any of a class of enzymes which catalyze the synthesis of polymeric molecules, especially nucleic acid polymers, typically by encouraging the base pairing of free nucleotides with those of an existing complementary template strand. DNA polymerases and RNA polymerases are essential for DNA replication and transcription, respectively.
- polymerase chain reaction (PCR)
- Any of a wide variety of molecular biology methods involving the rapid production of millions or billions of copies of a specific DNA sequence, allowing scientists to selectively amplify fragments of a very small sample to a quantity large enough to study in detail. In its simplest form, PCR generally involves the incubation of a target DNA sample of known or unknown sequence with a reaction mixture consisting of oligonucleotide primers, a heat-stable DNA polymerase, and free deoxyribonucleotide triphosphates (dNTPs), all of which are supplied in excess. This mixture is then alternately heated and cooled to pre-determined temperatures for pre-determined lengths of time according to a specified pattern which is repeated for many cycles, typically in a thermal cycler which automatically controls the required temperature variations. In each cycle, the most basic of which includes a denaturation phase, annealing phase, and elongation phase, the copies synthesized in the previous cycle are used as templates for synthesis in the next cycle, causing a chain reaction that results in the exponential growth of the total number of copies in the reaction mixture. Amplification by PCR has become a standard technique in virtually all molecular biology laboratories.
- polymerization
- The formation of a polymer from its constituent monomers; the chemical reaction or series of reactions by which monomeric subunits are covalently linked together into a polymeric chain or branching aggregate; e.g. the polymerization of a nucleic acid chain by linking consecutive nucleotides, a reaction catalyzed by a polymerase enzyme.
- polymorphism
- 1. In genetics, the regular and simultaneous existence of two or more discontinuous alleles or genotypes in the same population where the frequency of each allele is greater than can be explained by recurrent mutation alone, typically occurring in more than 1 percent of the population's individuals.[3] An example is the different human blood types (A, B, AB, and O).[5]
- 2. In chemistry, the existence of the same substance in two or more different crystalline forms.[5]
- polypeptide
- A long, continuous, and unbranched polymeric chain of amino acid monomers linked by covalent peptide bonds, typically longer than a peptide. Proteins generally consist of one or more polypeptides folded or arranged in a biologically functional way.
- polyploid
- (of a cell or organism) Having more than two homologous copies of each chromosome; i.e. any ploidy level that is greater than diploid. Polyploidy may occur as a normal condition of chromosomes in certain cells or even entire organisms, or it may result from errors in cell division or mutations causing the duplication of the entire chromosome set.
- polyribosome
- See polysome.
- polysaccharide
- A linear or branched polymeric chain of carbohydrate monomers (monosaccharides). Examples include glycogen and cellulose.[4]
- polysome
- A functional unit of protein synthesis consisting of multiple ribosomes attached along the length of the same messenger RNA transcript.[3]
- polysomy
- The condition of a cell or organism having at least one more copy of a particular chromosome than is normal for its ploidy level, e.g. a diploid organism with three copies of a given chromosome is said to show trisomy. Every polysomy is a type of aneuploidy.
- polytene chromosome
- position effect
- Any effect on the expression or functionality of a gene or sequence that is a consequence of its location or position within a chromosome or other DNA molecule. A sequence's precise location relative to other sequences and structures tends to strongly influence its activity and other properties, because different loci on the same molecule can have substantially different genetic backgrounds and physical/chemical environments, which may also change over time. For example, the transcription of a gene located very close to a nucleosome, centromere, or telomere is often repressed or entirely prevented because the proteins that make up these structures block access to the DNA by transcription factors, while the same gene is transcribed at a much higher rate when located in euchromatin. Proximity to promoters, enhancers, and other regulatory elements, as well as to regions of frequent transposition by mobile elements, can also directly affect expression; being located near the end of a chromosomal arm or to common crossover points may affect when replication occurs and the likelihood of recombination. Position effects are a major focus of research in the field of epigenetic inheritance.
- positional cloning
- A strategy for identifying and cloning a candidate gene based on knowledge of its locus or position alone and with little or no information about its products or function, in contrast to functional cloning. This method usually begins by comparing the genomes of individuals expressing a phenotype of unknown provenance (often a hereditary disease) and identifying genetic markers shared between them. Regions defined by markers flanking one or more genes of interest are cloned, and the genes located between the markers can then be identified by any of a variety of means, e.g. by sequencing the region and looking for open reading frames, by comparing the sequence and expression patterns of the region in mutant and wild-type individuals, or by testing the ability of the putative gene to rescue a mutant phenotype.[13]
- positive (+) sense strand
- See coding strand.
- positive control
- The initiation, activation, or enhancement of some biological process by the presence of a specific molecular entity (e.g. an activator or inducer), in the absence of which the process cannot proceed or is otherwise diminished.[5] In gene regulation, for example, the binding of an activating molecule such as a transcription factor to a promoter may recruit RNA polymerase to a coding sequence, thereby causing it to be transcribed. Contrast negative control.
- positive supercoiling
- The supercoiling of a double-stranded DNA molecule in the same direction as the turn of the double helix itself (e.g. a right-handed coiling of a helix with a right-handed turn).[9] Contrast negative supercoiling.
- post-transcriptional modification
- post-translational modification
- potency
- precursor cell
- A partially differentiated or intermediate stem cell with the ability to further differentiate into only one cell type; i.e. a unipotent stem cell that is the immediate parent cell from which fully differentiated cell types divide. The term "precursor cell" is sometimes used interchangeably with progenitor cell, though this term may also be considered technically distinct.
- Pribnow box
- primary structure
- primary transcript
- The unprocessed, single-stranded RNA molecule produced by the transcription of a DNA sequence as it exists before post-transcriptional modifications such as alternative splicing convert it into a mature RNA product such as an mRNA, tRNA, or rRNA. A precursor mRNA or pre-mRNA, for example, is a primary transcript which, after processing, becomes a mature mRNA ready for translation.
- primase
- Any of a class of enzymes that catalyze the synthesis of short, ~10-base RNA oligonucleotides, which by complementing the lagging strand during DNA replication are used as primers by DNA polymerase to initiate the synthesis of Okazaki fragments.[3]
- primer
- A short, single-stranded oligonucleotide, typically 5–100 bases in length, which "primes" or initiates nucleic acid synthesis by hybridizing to a complementary sequence on a template strand and thereby providing an existing 3'-end from which a polymerase can extend the new strand. Natural systems exclusively use RNA primers to initiate DNA replication and some forms of prokaryotic transcription, whereas the in vitro syntheses performed in many laboratory techniques such as PCR often use DNA primers. In modern laboratories, primers are carefully designed, often in "forward" and "reverse" pairs, to complement specific and unique sequences in target DNA molecules, with consideration given to their melting and annealing temperatures, and then purchased from commercial suppliers which create oligonucleotides on demand by de novo synthesis.
- primer dimer (PD)
- primer walking
- priming
- The initiation of nucleic acid synthesis by the hybridization or annealing of one or more primers to a complementary sequence within a template strand.
- pro-enzyme
- See zymogen.
- pro-protein
- An inactive precursor of a protein or polypeptide that is converted into the active form by some post-translational modification, such as by cleaving a specific peptide sequence from the precursor or by attaching other molecules to specific amino acid residues. The names of protein precursors are often prefixed with pro-, as in proinsulin. Enzyme precursors may be called pro-enzymes or zymogens.
- probe
- Any reagent used to make a single measurement in a biochemical assay such as a gene expression experiment. Molecules which have a specific affinity for one or more other molecules may be used to probe for the presence of those other molecules in samples of unknown composition. Probes are often labelled or otherwise used as reporters to indicate whether or not a specific chemical reaction is taking place. See also hybridization probe.
- probe-set
- A collection of two or more probes designed to measure a single molecular species, such as a collection of oligonucleotides designed to hybridize to various parts of the mRNA transcripts generated from a single gene.
- process molecular gene concept
- An alternative definition of a gene which emphasizes the contribution of non-DNA factors to the process by which the information encoded in a DNA sequence results in the synthesis of a polypeptide.
- prometaphase
- The second stage of cell division in mitosis, following prophase and preceding metaphase, during which the nuclear membrane disintegrates, the chromosomes inside form kinetochores around their centromeres, microtubules emerging from the poles of the mitotic spindle reach the nuclear space and attach to the kinetochores, and motor proteins associated with the microtubules begin to push the chromosomes toward the center of the cell.
- promoter
- A sequence or region of DNA, usually 100–1,000 base pairs long, which regulates the expression of one or more associated genes by containing binding sites for transcription factors which recruit RNA polymerase to the sequence and initiate transcription. Promoters are typically located immediately upstream of the genes they regulate, near to and often including the transcription start site.
- promotion
- See upregulation.
- prophase
- The first stage of cell division in both mitosis and meiosis, occurring after interphase and before prometaphase, during which the DNA of the chromosomes is condensed into chromatin, the nucleolus disintegrates, centrosomes move to opposite ends of the cell, and the mitotic spindle forms.
- protease
- Any of a class of enzymes which catalyze proteolysis, i.e. the decomposition of proteins into smaller polypeptides or individual amino acids, by cleaving peptide bonds via hydrolysis. Proteases are ubiquitous components of numerous biological pathways, and therefore it is often necessary to inhibit them in order for laboratory techniques involving protein activity to be effective.
- proteasome
- A large complex of protease enzymes that selectively degrades intracellular proteins which have been tagged for degradation by ubiquitination. Proteasomes play important roles in the timing and onset of cellular processes through the signal-mediated proteolysis of certain enzymes and regulatory proteins; they also contribute to the stress response by removing abnormal proteins and to the immune response by generating antigenic peptides.[5]
- protein
- A polymeric macromolecule composed of one or more long chains of amino acids linked by peptide bonds. Proteins are the three-dimensional structures created when these chains fold into specific higher-order arrangements following translation, and it is this folded structure which determines a protein's chemical activity and hence its biological function. Ubiquitous and fundamental in all living organisms, proteins are the primary means by which the activities of life are performed, participating in the vast majority of the biochemical reactions that occur inside and outside of cells. They are often classified according to the type(s) of reaction(s) they facilitate or catalyze, by the chemical substrate(s) they act upon, or by their functional role in cellular activity; e.g. as structural proteins, motor proteins, enzymes, transcription factors, or links within biochemical pathways.
- protein complex
- An assembly or aggregate of multiple proteins held together by intermolecular forces, especially one with a particular biological function. Complexes may include many of the same protein or all different proteins. Numerous cellular activities, including DNA replication, transcription, and translation, rely on protein complexes.[4]
- protein folding
- The physical process by which the linear chains of amino acids (i.e. polypeptides) synthesized during translation are changed from random coils into stable, orderly, three-dimensional shapes (i.e. proteins) by assuming a higher-order structure or conformation which permits the protein to be biologically functional, known as its native state. Folding is the consequence of amino acid residues participating in intermolecular electrostatic interactions with each other and with their surroundings, including other molecules, and so is strongly influenced by the particularities of the local chemical environment. The time it takes to properly fold a protein can vary greatly, but the process often begins while chain synthesis is still ongoing. Some chains may have motifs or domains which lack intrinsic order and remain unfolded across a wide range of chemical conditions. Having the correct three-dimensional structure is essential for proper protein function, and misfolded proteins are generally biologically inactive, though mutant folds can occasionally modify functionality in useful ways.
- protein kinase
- Any of a class of enzymes which phosphorylate proteins by catalyzing the transfer of a phosphate from ATP to an amino acid residue and often causing a functionally relevant conformational change as a result. The great majority of protein kinases phosphorylate the hydroxyl side chains of either serine, threonine, or tyrosine, though other types also exist.[3] Separate classes of kinases phosphorylate non-protein molecules such as lipids and carbohydrates.
- protein purification
- protein sorting
- The set of biological mechanisms by which proteins are directed and transported to appropriate destinations within or outside of the cell. Proteins must often be routed into the interior of organelles, embedded within a membrane, or secreted into the extracellular environment in order to serve their functions, and information contained in the protein itself instructs this delivery process.[28] In eukaryotic cells, an expansive network of organelles and pathways is specialized to facilitate protein sorting, including the endoplasmic reticulum and the Golgi apparatus.
- protein tag
- protein targeting
- See protein sorting.
- protein-coding gene
- A gene containing a coding sequence which can be transcribed and translated to produce a protein, as opposed to an RNA gene, which produces non-coding RNA transcripts that are not translated into proteins but instead have functions in and of themselves.
- proteinogenic amino acid
- Any of the 20 canonical amino acids which are encoded by the standard genetic code and incorporated into peptides and ultimately proteins during translation. The term may also be inclusive of an additional two amino acids encoded by non-standard codes which can be incorporated by special translation mechanisms.
- protein–protein interaction (PPI)
- proteoglycan
- Any heavily glycosylated protein, i.e. a core polypeptide with one or more covalently attached glycosaminoglycan chains. Proteoglycans are therefore considered a subclass of glycoproteins in which the carbohydrate units are long, linear polysaccharide polymers containing amino sugars[29] and generally bearing a net negative charge under physiological conditions due to the presence of sulfates and uronic acid groups. They are a major component of the extracellular matrix between animal cells, where they form large hydrated complexes commonly employed in connective tissues such as cartilage.
- proteolysis
- The decomposition of proteins into their component polypeptides or individual amino acids by cleaving the peptide bonds linking the amino acids together via hydrolysis. Proteolysis is an important reaction used not only for degrading and inactivating proteins but sometimes also to activate them by removing amino acid residues which inhibit their activity.[3] It is usually catalyzed by enzymes known as proteases.
- proteome
- The entire set of proteins that is or can be expressed by a particular genome, cell, tissue, or species at a particular time (such as during a single lifespan or during a specific developmental stage) or under particular conditions (such as when compromised by a certain disease).
- proteomics
- The study of the proteome of a particular genome, cell, or organism, i.e. the sum total of all of the proteins produced from it by translation. Proteomics technologies allow scientists to purify and identify proteins and polypeptides and determine which ones are most and least abundant at a given time or under a given experimental condition.
- protomer
- Any molecular subunit from which a larger polymeric macromolecule is built, including those subunits which are not strictly monomers and can themselves be divided into subunits. For example, a heterodimer of tubulin proteins is the protomer for microtubule assembly.[3]
- proton motive force
- See chemiosmotic coupling.
- protoplasm
- The biological contents enclosed within a membrane-bound space, variously referring to the cytoplasm, or the cytoplasm and nucleoplasm considered collectively, and sometimes exclusive of vacuoles.
- protoplast
- A plant, fungal, or bacterial cell which has had its cell wall removed by mechanical, chemical, or enzymatic means; or the complete contents (the protoplasm) of an intact cell excluding the cell wall.
- pulsatile secretion
- The secretion of substances from a cell, organelle, or tissue in a regular, rhythmic, pulse-like pattern. Many intercellular signaling molecules such as hormones and neurotransmitters are released in this manner in order to maintain homeostasis or to sensitize target cells by stimulating their production of surface receptors.
- purine
- A double-ringed heterocyclic organic compound which, along with pyrimidine, is one of two molecules from which all nitrogenous bases (including the nucleobases used in DNA and RNA) are derived. Adenine (A) and guanine (G) are classified as purines. The letter R is sometimes used to indicate a generic purine; e.g. in a nucleotide sequence read, R may be used to indicate that either purine nucleobase, A or G, can be substituted at the indicated position.
- putative gene
- A specific nucleotide sequence suspected to be a functional gene based on the identification of its open reading frame. The gene is said to be "putative" in the sense that no function has yet been described for its products.
- pyknosis
- The irreversible condensation of chromatin inside the nucleus as the cell undergoes necrosis or apoptosis, resulting in a compact mass which stains strongly and is conspicuous under a microscope.[13] It is followed by karyorrhexis.
- pyrimidine
- A single-ringed heterocyclic organic compound which, along with purine, is one of two molecules from which all nitrogenous bases (including the nucleobases used in DNA and RNA) are derived. Cytosine (C), thymine (T), and uracil (U) are classified as pyrimidines. The letter Y is sometimes used to indicate a generic pyrimidine; e.g. in a nucleotide sequence read, Y may be used to indicate that either pyrimidine nucleobase – C, T, or U – can be substituted at the indicated position.
- pyrimidine dimer
- A type of molecular lesion caused by photochemical damage to DNA or RNA, whereby exposure to ultraviolet (UV) radiation induces the formation of covalent bonds between pyrimidine bases occupying adjacent positions in the same polynucleotide strand, which in turn may cause local conformational changes in secondary structure and prevent base pairing with the opposite strand. In DNA, the dimerization reaction occurs between neighboring thymine and cytosine residues (T−T, C−C, or T−C); it can also occur between cytosine and uracil residues in double-stranded RNA. Pyrimidine dimers are usually quickly corrected by nucleotide excision repair, but uncorrected lesions can inhibit or arrest polymerase activity during transcription or replication.
- pyruvic acid
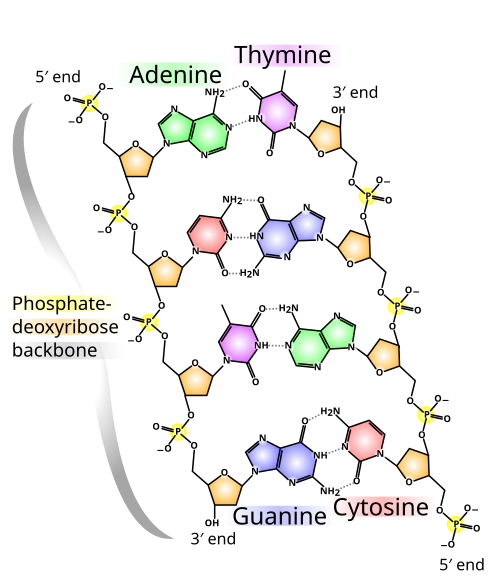
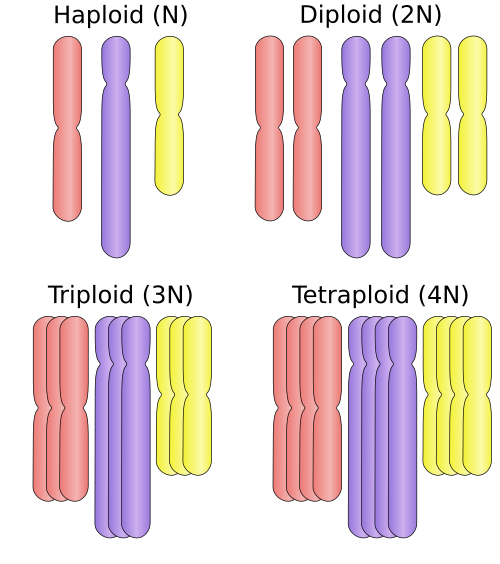

Q
[edit]- quantitative PCR (qPCR)
- quiescent culture
- A cell culture in which there is little or no active cell growth or replication but in which the cells nonetheless continue to survive, as observed with some confluent cultures.[2]
R
[edit]- random walk
- A popular description of the path followed by a locomotive cell or particle when there is no bias in movement, i.e. when the direction of movement at any given instant is not influenced by the direction of movement in the preceding instant. The essential randomness of cell movement in a uniform environment is only apparent over long periods of time, however; in the short term, cells can and do exhibit a tendency to continue moving in the same direction.[3]
- rDNA
- 1. An abbreviation of recombinant DNA.
- 2. An abbreviation of ribosomal DNA.
- reading frame
- A way of dividing the nucleotide sequence in a DNA or RNA molecule into a series of consecutive, non-overlapping groups of three nucleotides, known as triplets, which is how the sequence is interpreted or "read" by ribosomes during translation. In coding DNA, each triplet is referred to as a codon and corresponds to a particular amino acid to be added to the nascent peptide chain during translation. In general, only one reading frame (the so-called open reading frame) in a given sequence encodes a functional protein, though there are exceptions. A frameshift mutation results in a shift in the normal reading frame which affects all downstream codons and usually results in a completely different and senseless amino acid sequence.
- readthrough
- The continued transcription or translation of a DNA or mRNA sequence, respectively, beyond encoded termination signals such as a terminator sequence or a stop codon.[3]
- real-time PCR (rtPCR)
- See quantitative PCR.
- reassociation kinetics
- The measurement and manipulation of the rate of reannealing of complementary strands of DNA, generally by heating and denaturing a double-stranded molecule into single strands and then observing their rehybridization at a cooler temperature. Because the base pair G+C requires more energy to anneal than the base pair A+T, the rate of reannealing between two strands depends partly on their nucleotide sequence, and it is therefore possible to predict or estimate the sequence of the duplex molecule by the time it takes to fully hybridize. Reassociation kinetics is studied with C0t analysis: fragments reannealing at low C0t values tend to have highly repetitive sequences, while higher C0t values imply more unique sequences.[13]
- receptor
- A protein which initiates a cellular response to an external stimulus or propagates a molecular signal by binding a specific ligand, often a dedicated signaling molecule. Numerous types of receptors exist which serve an enormous variety of functions. Cell-surface receptors, such as those that bind acetylcholine and insulin, are embedded within the cell membrane with their binding sites exposed to the extracellular space; intracellular receptors, including many hormone receptors, are located in the cytoplasm, where they bind ligands that have diffused across the membrane and into the cell.[4]
- receptor-mediated endocytosis
- reciprocal translocation
- A type of chromosomal translocation by which there is a reciprocal exchange of chromosome segments between two or more non-homologous chromosomes. When the exchange of material is evenly balanced, reciprocal translocations are usually harmless.
- recognition site
- A specific motif or sequence, either of nucleotides in a nucleic acid molecule or of amino acids in a protein, that is "recognized" or identified by another protein in order to direct the protein's activity to a specific molecule or location. Recognition motifs may consist of a simple consecutive sequence within a single molecule or may involve multiple non-consecutive motifs, e.g. amino acids in separate parts of the same polypeptide which are brought into juxtaposition by the quaternary structure created during protein folding. Recognition sites often help to distinguish the nucleic acid or protein bearing the motif from other similar molecules and thereby identify it as a valid target for some biochemical activity, or to specify a locus or subregion within the larger macromolecule at which the activity is to occur. In this sense recognition sites are critical for properly localizing proteins to their biochemical targets. A protein's recognition site is often but not necessarily the same as its binding site or target site.[5]
- recombinant DNA (rDNA)
- Any DNA molecule in which laboratory methods of genetic recombination have brought together genetic material from multiple sources, thereby creating a sequence that would not otherwise be found in a naturally occurring genome. Because DNA molecules from all organisms share the same basic chemical structure and properties, DNA sequences from any species, or even sequences created de novo by artificial gene synthesis, may be incorporated into recombinant DNA molecules. Recombinant DNA technology is widely used in genetic engineering.
- recombinase
- Any of a class of enzymes united by their ability to catalyze some form of genetic recombination, by any of a variety of mechanisms, e.g. by site-specific recombination.
- recombination
- See genetic recombination, homologous recombination, and chromosomal crossover.
- recombinator
- Any nucleotide sequence that increases the likelihood of homologous recombination in nearby regions of the genome, e.g. the Chi sequence in certain species of bacteria.[13]
- recon
- The smallest unit of a DNA molecule capable of undergoing homologous recombination, i.e. a pair of consecutive nucleotides, adjacent to each other in cis.[13]
- regulon
- A group of non-contiguous genes which are regulated as a unit, generally by virtue of having their expression controlled by the same regulatory element or set of elements, e.g. the same repressor or activator. The term is most commonly used with prokaryotes, where a regulon may consist of genes from multiple operons.
- repeat
- Any pattern of nucleobases within a nucleic acid sequence which occurs in multiple copies in the same nucleic acid molecule such as a chromosome or within a genome. Repeated sequences are classified according to their length, structure, location, mode of replication, or evolutionary origin. They may be any length, but are often short motifs of less than 100 bases; they may be direct or inverted, and may occur in tandem arrays with the copies immediately adjacent to each other or interspersed with non-repeated sequences. Significant fractions of most eukaryotic genomes consist of repetitive DNA, much of it retroviral in origin, though repeats may also result from errors in normal cellular processes, as with duplications during DNA replication or cell division. Because so many genetic mechanisms depend on the binding or complementing of locally unique sequences, sequences containing or adjacent to repeats are particularly prone to errors in replication and transcription by strand slippage, or to forming problematic secondary structures, and thus repeats are often unstable in the sense that the number of copies tends to expand or diminish stochastically with each round of replication, causing great variation in copy number even between different cells in the same organism. When repeats occur within genes or regulatory elements, these properties often result in aberrant expression and lead to disease. Repeats are also essential for normal genome function in other contexts, as with telomeres and centromeres, which consist largely of repetitive sequences.
- repetitive DNA
- A region or fragment of DNA consisting largely or entirely of repeated nucleotide sequences.
- replacement mutation
- See nonsynonymous mutation.
- replication
- 1. The process by which certain biological molecules, notably the nucleic acids DNA and RNA, produce copies of themselves.
- 2. A technique used to estimate technical and biological variation in experiments for statistical analysis of microarray data. Replicates may be technical replicates, such as dye swaps or repeated array hybridizations, or biological replicates, biological samples from separate experiments which are used to test the effects of the same experimental treatment.
- replication eye
- The eye-shaped structure that forms when a pair of replication forks, each growing away from the origin, separates the strands of the double helix during DNA replication.
- replication fork
- The point at which the paired strands of a double-stranded DNA molecule are separated by helicase during DNA replication, breaking the hydrogen bonds between the complementary strands and thereby forming a structure with two branching single strands of DNA. Once unpaired, these strands serve as templates from which DNA polymerase synthesizes the leading strand and lagging strand. As replication proceeds, helicase moves along the DNA and continues to separate the strands, causing the replication fork to move as well.[3] A pair of replication forks forms when helicases work in opposite directions from a single origin of replication, creating a replication eye.
- replication rate
- The speed at which deoxyribonucleotides are incorporated into an elongating chain by DNA polymerases during DNA replication; or more generally the speed at which any chromosome, genome, cell, or organism makes a complete, independently functional copy of itself.
- replicator
- 1. Any fragment or region of DNA that contains a replication origin.[13]
- 2. Any molecule or structure capable of copying itself; namely, nucleic acids, but also crystals of many minerals, e.g. kaolinite.
- replicon
- Any molecule or region of DNA or RNA that replicates from a single origin of replication.
- replisome
- The entire complex of molecular machinery that carries out the process of DNA replication, including all proteins, nucleic acids, and other molecules which participate at an active replication fork.
- reporter
- In genetic engineering, a gene which when properly expressed encodes a gene product that is easily detected or visualized with biochemical assays (e.g. green fluorescent protein, β-galactosidase, chloramphenicol O-acetyltransferase, etc.), allowing researchers to use its expression in order to study the functions and properties of associated regulatory sequences. Reporters are commonly cloned into plasmid vectors in proximity to putative promoters, enhancers, or response elements, which are then mutated in order to precisely identify the specific recognition motifs within these sequences that are necessary for expression. In the broadest sense, reporters may also include things like molecular tags, fluorescent labels, and hybridization probes which render their conjugated molecules conspicuous or able to be purified; or they may be used similarly to selectable markers, to distinguish cells that express a given product from those that do not, so that researchers can easily identify mutants of interest or verify the success of an experimental treatment or laboratory procedure.[5]
- repression
- See downregulation.
- repressor
- A DNA-binding protein that inhibits the expression of one or more genes by binding to the operator and blocking the attachment of RNA polymerase to the promoter, thus preventing transcription. This process is known as negative gene regulation or repression.
- rescue
- The restoration of a defective cell or tissue to a healthy or normal condition,[13] or the reversion or recovery of a mutant gene to its normal functionality, especially in the context of experimental genetics, where an experiment (e.g. a drug, cross, or gene transfer) resulting in such a restoration is said to rescue the normal phenotype.
- residue
- An individual monomer or subunit of a larger polymeric macromolecule; e.g. a nucleic acid is composed of nucleotide residues, and a peptide or protein is composed of amino acid residues.[13]
- response element
- Any short sequence of DNA that serves some function related to the activity of a protein or other biomolecule, especially a sequence within a promoter region that is able to bind specific transcription factors in order to regulate transcription of specific genes.
- restitution
- The spontaneous rejoining of an experimentally broken chromosome which restores the original configuration.
- restitution nucleus
- A nucleus containing twice the expected number of chromosomes owing to an error in cell division, especially an unreduced, diploid product of meiosis resulting from the failure of the first or second meiotic division.
- restriction cloning
- The use of restriction sites and the enzymes specific for them in order to clone recombinant DNA molecules.
- restriction enzyme
- An endonuclease or exonuclease enzyme that recognizes and cleaves a nucleic acid molecule into fragments at or near specific recognition sequences known as restriction sites by breaking the phosphodiester bonds of the nucleic acid backbone. Restriction enzymes are naturally occurring in many organisms, but are also routinely used for artificial modification of DNA in laboratory techniques such as restriction cloning.
- restriction fragment
- Any DNA fragment that results from the cutting of a DNA strand by a restriction enzyme at one or more restriction sites.
- restriction fragment length polymorphism (RFLP)
- Variability within a population of organisms observed in the size of the restriction fragments produced when genomic DNA (or any particular DNA molecule) is digested by one or more restriction endonucleases. This variability results from a corresponding polymorphism in the locations of restriction sites within the molecule(s) due to slight differences in nucleotide sequence between individuals. RFLP is frequently exploited in the laboratory to construct physical maps of the genome, to identify the specific locus occupied by a particular gene, and to detect genetic differences between closely related individuals or determine that different samples originated from the same individual. Analysis of restriction fragments can also reveal the presence of a mutation that may itself cause disease or be closely linked to one that does.[5]
- restriction map
- A diagram of known restriction sites within a known DNA sequence, such as a plasmid vector, obtained by systematically exposing the sequence to various restriction enzymes and then comparing the lengths of the resulting fragments, a technique known as restriction mapping. See also gene map.
- restriction mapping
- The use of type II restriction endonucleases to cleave DNA molecules at specific restriction sites in order to produce characteristic patterns of fragments which can be resolved by size using gel electrophoresis. Digesting DNA molecules such as genomic DNA or plasmids with one or multiple restriction enzymes makes it possible to deduce from the sizes of the resulting fragments the order or arrangement of the restriction sites within the molecule and the distances between them, and thus to construct reliable maps with restriction sites effectively serving as genetic markers.[5]
- restriction site
- A short, specific sequence of nucleotides (typically 4 to 8 bases in length) that is reliably recognized by a particular restriction enzyme. Because restriction enzymes usually bind as homodimers, restriction sites are generally palindromic sequences spanning both strands of a double-stranded DNA molecule. Restriction endonucleases cleave the phosphate backbone between two nucleotides within the recognized sequence itself, while other types of restriction enzymes make their cuts at one end of the sequence or at a nearby sequence.
- retrogene
- A gene or other DNA sequence which has arisen in a genome by the stable integration of the genetic material of a retrovirus, e.g. by the reverse transcription of viral RNA and the subsequent insertion of the resulting DNA fragments into the host cell's genomic DNA.[5] These endogenous viral elements can then be replicated along with the host's own genes and thus persist indefinitely in the host genome, and in many cases retain the ability to produce functional viral proteins from this latent state, by which they may continue to copy, excise, and/or transpose themselves.
- reverse genetics
- An experimental approach in molecular genetics in which a researcher starts with a known gene and attempts to determine its function or its effect on phenotype by any of a variety of laboratory techniques, commonly by deliberately mutating the gene's DNA sequence or by repressing or silencing its expression and then screening the mutant organisms for changes in phenotype. When the gene of interest is the only one in the genome whose expression has been manipulated, any observed phenotypic changes are assumed to be influenced by it. This is the opposite of forward genetics, in which a known phenotype is linked to one or more unknown genes.
- reverse transcriptase (RT)
- An enzyme capable of synthesizing a complementary DNA molecule from an RNA template, a process termed reverse transcription.
- reverse transcription
- The synthesis of a DNA molecule from an RNA template, i.e. the opposite of ordinary transcription. This process, mediated by an enzyme known as a reverse transcriptase, is used by many viruses to replicate their genomes, as well as by retrotransposons and in some eukaryotic cell types.
- ribonuclease (RNase)
- Any of a class of nuclease enzymes which catalyze the hydrolytic cleavage of phosphodiester bonds in RNA molecules, thus severing polymeric strands of ribonucleotides into smaller components. Compare deoxyribonuclease.
- ribonucleic acid (RNA)
- A polymeric nucleic acid molecule composed of a series of ribonucleotides which incorporate a set of four nucleobases: adenine (A), guanine (G), cytosine (C), and uracil (U). Unlike DNA, RNA is more often found as a single strand folded onto itself, rather than a paired double strand. Various types of RNA molecules serve in a wide variety of essential biological roles, including coding, decoding, regulating, and expressing genes, as well as functioning as signaling molecules and, in certain viral genomes, as the primary genetic material itself.
- ribonucleoprotein (RNP)
- A nucleoprotein that is a complex of one or more RNA molecules and one or more proteins. Examples include ribosomes and the enzyme ribonuclease P.
- ribonucleotide
- A nucleotide containing ribose as its pentose sugar component, and the monomeric subunit of ribonucleic acid (RNA) molecules. Ribonucleotides canonically incorporate any of four nitrogenous bases: adenine (A), guanine (G), cytosine (C), and uracil (U). Compare deoxyribonucleotide.
- ribonucleotide reductase (RNR)
- An enzyme which catalyzes the formation of deoxyribonucleotides via the reductive dehydroxylation of ribonucleotides, specifically by removing the 2' hydroxyl group from the ribose ring of ribonucleoside diphosphates (rNDPs). RNR plays a critical role in regulating the overall rate of DNA synthesis such that the ratio of DNA to cell mass is kept constant during cell division and DNA repair.
- ribose
- A monosaccharide sugar which, as D-ribose in its pentose ring form, is one of three primary components of the ribonucleotides from which ribonucleic acid (RNA) molecules are built. Ribose differs from its structural analog deoxyribose only at the 2' carbon, where ribose has an attached hydroxyl group that deoxyribose lacks.
- ribosomal DNA (rDNA)
- A DNA sequence that codes for ribosomal RNA (rRNA). In many eukaryotic genomes, rDNA occupies large, highly conserved regions of multiple chromosomes and is rich in both genes and repeats.
- ribosomal RNA (rRNA)
- A type of non-coding RNA which is the primary constituent of ribosomes, binding to ribosomal proteins to form the small and large subunits. It is ribosomal RNA which enables ribosomes to perform protein synthesis by working as a ribozyme that catalyzes the set of reactions comprising translation. Ribosomal RNA is transcribed from the corresponding ribosomal DNA (rDNA) and is the most abundant class of RNA in most cells, bearing responsibility for the translation of all encoded proteins despite never being translated itself.
- ribosome
- A macromolecular complex made of both RNA and protein which serves as the site of protein synthesis by translation. Ribosomes have two subunits, each of which consists of one or more strands of ribosomal RNA bound to various ribosomal proteins: the small subunit, which reads the messages encoded in messenger RNA molecules, and the large subunit, which links amino acids in sequence to form a polypeptide chain. Ribosomes are essential and ubiquitous in all cell types and are used by all known forms of life.
- riboswitch
- A regulatory sequence within a messenger RNA transcript that can bind a small effector molecule, preventing or disrupting translation and thereby acting as a switch that regulates the mRNA's expression.
- ribozyme
- An RNA molecule with enzymatic activity,[4] i.e. one that is capable of catalyzing one or more specific biochemical reactions, similar to protein enzymes. Ribozymes function in numerous capacities, including in ribosomes as part of the large subunit ribosomal RNA.
- RNA
- See ribonucleic acid.
- RNA gene
- A gene that codes for any of the various types of non-coding RNA (e.g. rRNA and tRNA).[13]
- RNA interference (RNAi)
- RNA polymerase
- Any of a class of polymerase enzymes that synthesize RNA molecules from a DNA template. RNA polymerases are essential for transcription and are found in all living organisms and many viruses. They build long single-stranded polymers called transcripts by adding ribonucleotides one at a time in the 5'-to-3' direction, relying on the template provided by the complementary strand to transcribe the nucleotide sequence faithfully. Unlike DNA polymerases, RNA polymerases notably do not require oligonucleotide primers to initiate synthesis; i.e. they are capable of synthesizing RNA molecules de novo.
- RNA splicing
- RNA-induced silencing complex (RISC)
- A ribonucleoprotein complex which works to silence endogenous and exogenous genes by participating in various RNA interference pathways at the transcriptional and translational levels. RISC can bind both single-stranded and double-stranded RNA fragments and then cleave them or use them as guides to target complementary mRNAs for degradation.
- RNase
- See ribonuclease.
- Robertsonian translocation (ROB)
- A type of chromosomal translocation by which double-strand breaks at or near the centromeres of two acrocentric chromosomes cause a reciprocal exchange of segments that gives rise to one large metacentric chromosome (composed of the long arms) and one extremely small chromosome (composed of the short arms), the latter of which is often subsequently lost from the cell with little effect because it contains very few genes. The resulting karyotype shows one fewer than the expected total number of chromosomes, because two previously distinct chromosomes have essentially fused together. Carriers of Robertsonian translocations are generally not associated with any phenotypic abnormalities, but do have an increased risk of generating meiotically unbalanced gametes.
- rolling circle replication (RCR)
- rough endoplasmic reticulum (RER)
- A type of membrane in the endoplasmic reticulum with numerous ribosomes conspicuously attached to its surface, in contrast to the "smooth" endoplasmic reticulum which lacks ribosomes. The rough ER serves as the site of protein synthesis for the majority of the cell's secreted and transmembrane proteins, as well as the site of synthesis of membrane lipids. It may be continuous with the smooth ER or exist separately.[3]
- rRNA
- See ribosomal RNA.
- rtPCR
- 1. An abbreviation of real-time polymerase chain reaction, synonymous with quantitative PCR.
- 2. An abbreviation of reverse transcription polymerase chain reaction.


S
[edit]- S phase
- The phase of the cell cycle during which nuclear DNA is replicated, occurring after the G1 phase and before the G2 phase.[2]
- salvage pathway
- Any metabolic pathway that utilizes compounds formed in catabolism for the anabolism or biosynthesis of new compounds, e.g. by recycling building block monomers such as free purine and pyrimidine bases to make new nucleotides.[5]
- samesense mutation
- See synonymous mutation.
- Sanger sequencing
- A method of DNA sequencing based on the in vitro replication of a DNA template sequence, during which fluorochrome-labeled, chain-terminating dideoxynucleotides are randomly incorporated in the elongating strand; the resulting fragments are then sorted by size with electrophoresis, and the particular fluorochrome terminating each of the size-sorted fragments is detected by laser chromatography, thus revealing the sequence of the original DNA template through the order of the fluorochrome labels as one reads from small-sized fragments to large-sized fragments. Though Sanger sequencing has been replaced in some contexts by next-generation methods, it remains widely used for its ability to produce relatively long sequence reads (500+ nucleotides) and its very low error rate.
- saturation hybridization
- An in vitro nucleic acid hybridization reaction in which one polynucleotide component (either DNA or RNA) is supplied in great excess relative to the other, causing all complementary sequences in the other polynucleotide to pair with the excess sequences and form hybrid duplex molecules.[13]
- scRNA
- See small conditional RNA.
- second messenger
- A molecule or compound (often a protein) that is caused to accumulate in an effector cell by the action of a hormone, growth factor, or other agonist and thereby brings about the action of that agonist on the cell. Second messengers are therefore critical mediators of a diverse variety of signal transduction pathways, including the synthesis of cyclic AMP by adenylate cyclase and of cyclic GMP by guanylate cyclase, the opening of ion channels, and the phosphorylation of proteins by serine/threonine-specific or tyrosine-specific protein kinases.[5]
- secondary structure
- The arrangement or folding of a polypeptide's primary structure into higher-order, locally organized structures, primarily via hydrogen bonding between non-adjacent amino acid residues, in particular α helices and β sheets; or the arrangement of double-stranded nucleic acid chains into the shape of a double helix stabilized by hydrogen bonds between the complementary bases.[5]
- second-generation sequencing
- See massively parallel sequencing.
- selectable marker
- A gene or other genetic material whose expression in cultured cells confers a selective advantage in the culture environment, causing cells expressing the gene to have one or more traits suitable for artificial selection. Selectable markers are widely used in the laboratory as a type of reporter, usually to indicate the success of a procedure meant to introduce exogenous DNA into a host cell such as transfection or transformation. A common example is an antibiotic resistance gene which is transformed into competent bacterial cells cultured on a medium containing the particular antibiotic, such that only those cells which have successfully taken up and expressed the gene are able to survive and grow into colonies.
- selfish genetic element
- Any genetic material (e.g. a gene or any other DNA sequence) which can enhance its own replication and/or transmission into subsequent generations at the expense of other genes in the genome, even if doing so has no positive effect or even a net negative effect on the fitness of the genome as a whole. Selfish elements usually work by producing self-acting gene products which repeatedly copy and paste their own coding sequences into other parts of the genome, independently of normal DNA replication (as with transposable elements); by facilitating the uneven swapping of chromosome segments during genetic recombination events (as with unequal crossing over); or by disrupting the normally equal redistribution of replicated material during mitosis or meiosis such that the probability that the selfish element is present in a given daughter cell is greater than the normal 50 percent (as with gene drives).
- semiconservative replication
- The standard mode of DNA replication that occurs in all living cells, in which each of the two parental strands of the original double-stranded DNA molecule are used as template strands, with DNA polymerases replicating each strand separately and simultaneously in antiparallel directions. The result is that each of the two double-stranded daughter molecules is composed of one of the original parental strands and one newly synthesized complementary strand, such that each daughter molecule conserves the precise sequence of information (indeed the very same atoms) from one-half of the original molecule. Contrast conservative replication and dispersive replication.
- sense
- A distinction made between the individual strands of a double-stranded DNA molecule in order to easily and specifically identify each strand. The two complementary strands are distinguished as sense and antisense or, equivalently, the coding strand and the template strand. It is the antisense/template strand which is actually used as the template for transcription; the sense/coding strand merely resembles the sequence of codons on the RNA transcript, which makes it possible to determine from the DNA sequence alone the expected amino acid sequence of any protein translated from the RNA transcript. Which strand is which is relative only to a particular RNA transcript and not to the entire DNA molecule; that is, either strand can function as the sense/coding or antisense/template strand.
- sense codon
- Any codon that specifies an amino acid, as opposed to a stop codon, which does not specify any particular amino acid but instead signals the end of translation.
- sequence
- See nucleic acid sequence.
- sequence logo
- In bioinformatics, a graphical representation of the conservation of nucleobases or amino acids at each position within a nucleic acid or protein sequence. Sequence logos are created by aligning many sequences and used to depict consensus sequences as well as the degree of variability within the pool of aligned sequences.
- sequence-tagged site (STS)
- Any DNA sequence that occurs exactly once within a particular genome, and whose location and nucleotide sequence are known with confidence.
- sequencing
- The determination of the order or sequence of nucleotides in a nucleic acid molecule, or of amino acids in a peptide, by any means. Sequences are usually written as a linear string of letters which conveniently summarizes much of the atomic-level structure of the molecule.
- sex chromosome
- See allosome.
- sex linkage
- The presence of a particular gene or DNA sequence on a sex chromosome (in mammals either the X chromosome or the Y chromosome) rather than on an autosome; these genes are said to be sex-linked. Expression of sex-linked genes varies by organism depending on the mechanism of sex determination and the types of sex chromosomes present, but the associated phenotypes often exclusively appear in either the homogametic or heterogametic sex.[2]
- Shine–Dalgarno sequence
- In many prokaryotic messenger RNAs, the consensus sequence AGGAGGU, located 6–8 bases upstream of the translation start codon, which functions as a binding site for the ribosome by complementing a sequence in the ribosomal RNA.[13]
- short arm
- In condensed chromosomes where the positioning of the centromere creates two segments or "arms" of unequal length, the shorter of the two arms of a chromatid. Contrast long arm.
- short interspersed nuclear element (SINE)
- short tandem repeat (STR)
- See microsatellite.
- shotgun sequencing
- A method of sequencing entire genomes in which genomic DNA is randomly fragmented (e.g. by sonication or restriction digests), cloned into plasmid vectors, and then sequenced using primers that anneal to flanking sequences in the plasmids. Computer software is used to align the sequenced fragments via overlapping contigs,[5] allowing scientists to deduce the relative genomic locations of each fragment and thereby assemble a complete genome.
- signal peptidase
- signal peptide
- Any sequence of amino acids, usually 15–30 residues in length, that functions as a molecular signal directing the sorting and transport of the polypeptide bearing it to a specific location within a cell or organelle. Signal peptides are commonly located close to either the N-terminal or C-terminal ends of nascent or recently synthesized polypeptides, especially those destined for secretion from the cell or integration into a membrane, and are typically cleaved off by a dedicated signal peptidase when their polypeptide reaches the endoplasmic reticulum.[5] Similar but distinct varieties of protein targeting are accomplished with nuclear localization signals and post-translational protein tags.
- signal transduction
- The process by which a chemical, electrical, or mechanical signal is converted into a cellular response, or the transmission or propagation of such a signal through a cell as a series of molecular events known as a signaling pathway. For example, the extracellular interaction of a hormone, growth factor, or some other chemical agonist with a specific cell surface receptor can trigger a cascade of sequential biochemical reactions which propagate through the cell membrane and into the cytoplasm, provoking the synthesis of second messengers and leading to amplification of the signal or activation of other pathways. Other modes of transduction involve agonists which diffuse across the membrane freely, eliciting intracellular changes without amplification, or rapid shifts in cell polarity which transmit electrical impulses, such as those that cause the axons of neural cells to release neurotransmitters at synapses.[5]
- silencer
- A sequence or region of DNA that can be bound by a repressor, thereby blocking the transcription of a nearby gene.
- silencing
- The total or near-total loss of expression of a particular gene or DNA sequence by any mechanism, natural or artificial, whether before, during, or after transcription or translation, which completely prevents the normal gene product from being produced and thereby deprives the cell of its ordinary function. Gene silencing may occur via natural regulatory mechanisms such as condensation of the relevant segment of DNA into a transcriptionally inactive, heterochromatic state, in which case the term is more or less equivalent to repression; genes are also commonly silenced artificially for research purposes by using techniques such as knockdown (e.g. by RNA interference) or knockout (by deleting the gene from the genome entirely). See also downregulation.
- silent allele
- An allele that does not produce a detectable product.[13] Compare null allele.
- silent mutation
- A type of neutral mutation which does not have an observable effect on the organism's phenotype. Though the term "silent mutation" is often used interchangeably with synonymous mutation, synonymous mutations are not always silent, nor vice versa. Missense mutations which result in a different amino acid but one with similar functionality (e.g. leucine instead of isoleucine) are also often classified as silent, since such mutations usually do not significantly affect protein function.
- simple sequence repeat (SSR)
- See microsatellite.
- single-nucleotide polymorphism (SNP)
- Any substitution of a single nucleotide which occurs at a specific position within a genome and with measurable frequency within a population; for example, at a specific base position in a DNA sequence, the majority of the individuals in a population may have a cytosine (C), while in a minority of individuals, the same position may be occupied by an adenine (A). SNPs are usually defined with respect to a "standard" reference genome; an individual human genome differs from the reference human genome at an average of 4 to 5 million positions, most of which consist of SNPs and short indels. See also polymorphism.
- single-strand break (SSB)
- The loss of continuity of the phosphate-sugar backbone in one strand of a DNA duplex.[9] See also nick; contrast double-strand break.
- single-stranded
- Composed of a single, unpaired nucleic acid molecule, i.e. one linear strand of nucleotides sharing a single phosphodiester backbone, as opposed to a duplex of two such strands joined by base pairing. See also single-stranded DNA and single-stranded RNA.
- single-stranded DNA (ssDNA)
- Any DNA molecule that consists of a single nucleotide polymer or strand, as opposed to a pair of complementary strands held together by hydrogen bonds (double-stranded DNA). In most circumstances, DNA is more stable and more common in double-stranded form, but high temperatures, low concentrations of dissolved salts, and very high or low pH can cause double-stranded molecules to decompose into two single-stranded molecules in a denaturation process known as melting; this reaction is exploited by naturally occurring enzymes such as those involved in DNA replication as well as by laboratory techniques such as polymerase chain reaction.
- siRNA
- See small interfering RNA.
- sister chromatids
- A pair of identical copies (chromatids) produced as the result of the DNA replication of a chromosome, particularly when both copies are joined together by a common centromere; the pair of sister chromatids is called a dyad. The two sister chromatids are ultimately separated from each other into two different cells during mitosis or meiosis.
- site-directed mutagenesis
- small conditional RNA (scRNA)
- A class of small RNA molecules engineered so as to change conformation conditionally in response to cognate molecular inputs, often with the goal of controlling signal transduction pathways in vitro or in vivo.
- small interfering RNA (siRNA)
- small nuclear RNA (snRNA)
- A class of small non-coding RNA molecules, approximately 100–300 nucleotides in length and rich in uridine residues, found in association with specific proteins as part of ribonucleoprotein complexes known as snRNPs within nuclear speckles and Cajal bodies of the eukaryotic nucleus.[5] SnRNPs assemble into larger complexes known as spliceosomes which play important roles in the splicing of pre-mRNA transcripts (hnRNAs) before they are exported to the cytoplasm.
- small nucleolar RNA (snoRNA)
- A class of small non-coding RNA molecules whose primary function is to direct the post-transcriptional modification of other RNAs, mainly transfer RNAs (tRNA), small nuclear RNAs (snRNA), and especially ribosomal RNAs (rRNA) as a part of ribosome synthesis in the nucleolus. SnoRNAs contain antisense sequences that complement sequences within these target RNAs and guide ribonucleoprotein complexes to them, which can then catalyze specific nucleoside modifications, typically methylation or pseudouridylation.
- small temporal RNA (stRNA)
- A subclass of microRNAs, originally described in nematodes, which regulate the timing of developmental events by binding to complementary sequences in the 3' untranslated regions of messenger RNAs and inhibiting their translation. In contrast to siRNAs, which serve similar purposes, stRNAs bind to their target mRNAs after the initiation of translation and without affecting mRNA stability, which makes it possible for the target mRNAs to resume translation at a later time.
- small ubiquitin-like modifier (SUMO)
- Any of a family of small proteins, each approximately 100 amino acids, which are covalently conjugated to and removed from charged residues of other proteins in a form of post-translational modification known as SUMOylation, thereby functioning as a protein tag in a manner resembling ubiquitin.
- smooth endoplasmic reticulum (SER)
- A type of membrane in the endoplasmic reticulum that lacks ribosomes on its surface, thus visibly contrasting with the "rough" endoplasmic reticulum. Smooth ER tends to be tubular rather than sheet-like, and may form an extension of the rough ER or exist separately. It is especially abundant in cells concerned with lipid metabolism.[3]
- snoRNA
- See small nucleolar RNA.
- snRNA
- See small nuclear RNA.
- soluble RNA (sRNA)
- See transfer RNA.
- somatic cell
- Any biological cell forming the body of an organism, or, in multicellular organisms, any cell other than a gamete, germ cell, or undifferentiated stem cell. Somatic cells are theoretically distinct from cells of the germ line, meaning the mutations they have undergone can never be transmitted to the organism's descendants, though in practice exceptions do exist.
- somatic cell nuclear transfer (SCNT)
- somatic crossover
- See mitotic recombination.
- sonication
- The application of sound energy at ultrasonic frequencies in order to agitate particles in a chemical or biological sample. Intense acoustic vibrations produce pressure waves and cavitations that propagate through a liquid medium, thereby converting the sound energy to mechanical energy which can disperse solutes, disrupt intermolecular interactions, and break covalent bonds. At various amplitudes sonication can be used to increase the permeability of cell or nuclear membranes, a technique known as sonoporation, or to completely destroy them and release their contents for isolation and extraction. It has a wide variety of applications in industry and research, including creating nanoparticles such as liposomes, shearing DNA and proteins into smaller fragments, degassing liquids, and ultrasonic cleaning.
- Southern blotting
- A molecular biology method used to detect a specific sequence in DNA samples. The method combines separation of DNA fragments by gel electrophoresis, transfer of the DNA to a synthetic membrane, and subsequent identification of target fragments with radio-labeled or fluorescent hybridization probes. Compare northern blotting, western blotting, and eastern blotting.
- spacer
- Any sequence or region of non-coding DNA separating neighboring genes, whether transcribed or not. The term is used in particular to refer to the non-coding regions between the many repeated copies of the ribosomal RNA genes.[9] See also intergenic region.
- spatially-restricted gene expression
- The expression of one or more genes only within a specific anatomical region or tissue, often in response to a paracrine signal. The boundary between the jurisdictions of two spatially restricted genes may generate a sharp phenotypic gradient there, as with striping patterns.
- spindle apparatus
- The cytoskeletal structure that forms during cell division in eukaryotic cells, consisting of a network of long, flexible microtubules extending from each pole of the parent cell and attaching to kinetochores at the centromeres of homologous chromosomes or sister chromatids near the cell equator. As the microtubules shorten, they pull the chromosomes apart, separating them into different daughter cells. Proper formation of this apparatus is critical in both mitosis and meiosis, where it is respectively referred to as the mitotic spindle and meiotic spindle.
- spliceosome
- A large ribonucleoprotein complex found primarily in the nucleus of eukaryotic cells, composed of multiple small nuclear ribonucleoproteins (snRNPs) which are themselves composed of small nuclear RNAs (snRNAs) and a variety of snRNP-specific proteins. Spliceosomes assemble at the junctions between exons and introns within primary RNA transcripts and catalyze the removal of the introns and the ligation of the flanking exons in a form of post-transcriptional processing known as RNA splicing.[30]
- splicing
- Any natural or artificial process involving the excision of oligonucleotide sequences from nucleic acid molecules (either DNA or RNA) or of peptide sequences from proteins and the subsequent re-ligation of the flanking fragments into a single continuous molecule lacking the excised sequence. RNA splicing in particular is an important form of post-transcriptional processing whereby introns are removed from primary mRNA transcripts and the exons rejoined (and sometimes rearranged) to create mature mRNAs; a similar process also occurs with the removal of inteins and the rejoining of exteins in the post-translational modification of certain proteins. The term may also refer more generally to artificial techniques for creating recombinant sequences in genetic engineering.[5]
- sRNA
- See transfer RNA.
- ssDNA
- See single-stranded DNA.
- ssRNA
- See single-stranded RNA.
- standard genetic code
- The genetic code used by the vast majority of living organisms for translating nucleic acid sequences into proteins. In this system, of the 64 possible permutations of three-letter codons that can be made from the four nucleotides, 61 code for one of the 20 amino acids, and the remaining three code for stop signals. For example, the codon CAG codes for the amino acid glutamine and the codon UAA is a stop codon. The standard genetic code is described as degenerate or redundant because some amino acids can be coded for by more than one different codon.
- start codon
- The first codon translated by a ribosome from a mature messenger RNA transcript, used as a signal to initiate peptide synthesis. In the standard genetic code, the start codon always codes for the same amino acid, methionine, in eukaryotes and for a modified methionine in prokaryotes. The most common start codon is the triplet AUG. Contrast stop codon.
- statistical genetics
- A branch of genetics concerned with the development of statistical methods for drawing inferences from genetic data. The theories and methodologies of statistical genetics often support research in quantitative genetics, genetic epidemiology, and bioinformatics.
- stem cell
- Any biological cell which has not yet differentiated into a specialized cell type and which can divide through mitosis to produce more undifferentiated stem cells.
- stem-loop
- See hairpin.
- sticky end
- A term used to describe the end of a double-stranded DNA molecule where one strand is longer than the other by one or more nucleobases, creating a single-stranded "overhang" of unpaired bases, in contrast to a so-called blunt end, where no such overhang exists because the terminal nucleobases on each strand are base-paired with each other. Blunt ends and sticky ends are relevant when ligating linear DNA molecules, e.g. in restriction cloning, because many restriction enzymes cleave the phosphate backbone in a way that leaves behind terminal overhangs in the digested fragments. These sticky-ended molecules ligate much more readily with other sticky-ended molecules having complementary overhangs, allowing scientists to ensure that specific DNA fragments are ligated together in specific places.
- stop codon
- A codon that signals the termination of protein synthesis during translation of a messenger RNA transcript. In the standard genetic code, three different stop codons are used to dissociate ribosomes from the growing amino acid chain, thereby ending translation: UAG (nicknamed "amber"), UAA ("ochre"), and UGA ("opal"). Contrast start codon.
- strand
- An individual chain of nucleotides comprising a nucleic acid polymer, existing either independently (in which case the nucleic acid molecule is said to be single-stranded) or base-paired in a duplex with another, separate strand (in which case it is said to be double-stranded).
- stringency
- The effect of conditions such as temperature and pH upon the degree of complementarity that is required for a hybridization reaction to occur between two single-stranded nucleic acid molecules. In the most stringent conditions, only exact complements can successfully hybridize; as stringency decreases, an increasing number of mismatches can be tolerated between the two hybridizing strands.[18]
- stRNA
- See small temporal RNA.
- structural gene
- A gene that codes for any protein or RNA product other than a regulatory factor. Structural gene products include enzymes, structural proteins, and certain non-coding RNAs.
- structural protein
- Any protein which contributes to the mechanical shape and structure of cells, organelles, or tissues (e.g. collagen and actin), as distinguished from proteins which serve some other purpose, such as enzymes. This distinction is not well-defined, however, as many proteins have both structural and non-structural roles.[2]
- subcellular localization
- 1. The subdivision of the interior of a cell into functionally distinct spaces or compartments (e.g. membrane-bound organelles) and the delegation of particular cellular functions and activities to these particular spaces.
- 2. The determination by any of various laboratory methods (e.g. fluorescent labelling) of the precise location(s) within a cell where a specific molecule has occupancy, or at which a specific activity occurs.
- submetacentric
- (of a linear chromosome or chromosome fragment) Having a centromere positioned close to but not exactly in the middle of the chromosome, resulting in chromatid arms of slightly different lengths.[6] Compare metacentric.
- substitution
- A type of point mutation in which a single nucleotide and its attached nucleobase is replaced by a different nucleotide.
- substrate
- 1. A chemical compound or molecule upon which a particular enzyme directly acts, often but not necessarily binding the molecule by forming one or more chemical bonds.[2] See also ligand.
- 2. The substance, biotic or abiotic, upon which an organism grows or lives, or by which it is supported; e.g. a particular growth medium used in cell culture. See also substratum.
- substratum
- A solid surface to which a cell or organism adheres or by which it is supported, or over which it moves.[3] See also substrate.
- subunit
- A single unit of a multi-unit compound or molecular aggregate; e.g. a monomer from which a larger polymer is composed (as with nucleotides in nucleic acids), or an individual polypeptide chain in a multi-chain protein, or an entire protein which participates alongside other proteins as part of a protein complex.[2][4]
- SUMOylation
- A type of post-translational modification in which a SUMO protein is conjugated to a polar residue of another protein (usually a lysine) via a covalent isopeptide bond. This effectively tags the second protein, making it distinguishable to other biomolecules and in many cases allowing it to participate in specific reactions or to interact with specific protein complexes. SUMOlyation is closely related to ubiquitination, relying on the same E1/E2/E3 enzymes to transfer SUMO to specific recognition motifs in the target protein, though detaching SUMO depends on SUMO-specific proteases. It plays important roles in numerous cellular processes, including protein localization, transcriptional regulation, stress-response pathways, and cell cycle checkpoints, among others. SUMOlyation is also used in the laboratory as a molecular label and to help solubilize proteins which are difficult to purify.
- supercoiling
- supersecondary structure
- suppression
- See downregulation.
- suspension culture
- A type of cell culture in which individual cells or aggregates of cells are suspended in a liquid growth medium, and usually prevented from settling by continuous gentle agitation. Many prokaryotic and eukaryotic cell types readily proliferate in suspension cultures, but they are particularly useful for culturing non-adherent cell lines such as hematopoietic cells, plant cells, and insect cells. Compare adherent culture.
- symporter
- Any of a class of transmembrane transporter proteins which facilitate the transport of two or more different molecules across the membrane at the same time and in the same direction; e.g. glucose and sodium ions. Contrast antiporter and uniporter.
- synapsis
- synaptonemal complex
- A complex of scaffolding proteins that mediates synapsis and homologous recombination between the chromatids of homologous chromosomes during prophase I of meiosis.
- syncytium
- A multinucleate cell, i.e. a cell containing more than one nucleus or, in the broadest sense, more than one nuclear genome (a meaning which is equated with polyploidy). Syncytia may form as a result of cell fusion between uninucleate cells, migration of a nucleus from one cell to another, or multiple nuclear divisions without accompanying cytokinesis (forming a coenocyte).[9] The term may also refer to cells which are interconnected by specialized membranes with gap junctions as in some neuromuscular cell types.
- syndesis
- The synapsis of chromosomes during meiosis.[13]
- synezis
- The aggregation of chromosomes into a dense knot that adheres to one side of the nucleus, commonly observed during leptonema in certain organisms.[13]
- synonymous mutation
- A type of mutation in which the substitution of one nucleotide base for another results, after transcription and translation, in an amino acid sequence which is identical to the original unmutated sequence. This is possible because of the degeneracy of the genetic code, which allows different codons to code for the same amino acid. Though synonymous mutations are often considered silent, this is not always the case; a synonymous mutation may affect the efficiency or accuracy of transcription, splicing, translation, or any other process by which genes are expressed, and thus become effectively non-silent. Contrast nonsynonymous mutation.
- synthesis phase
- See S phase.



T
[edit]- tandem repeat
- A pattern within a nucleic acid sequence in which one or more nucleobases are repeated and the repetitions are directly adjacent (i.e. tandem) to each other. An example is ATGACATGACATGAC, in which the sequence ATGAC is repeated three times.
- target site
- The site or locus upon another molecule at which a protein performs a particular biochemical activity; e.g. the nucleotide motif at which a restriction endonuclease cleaves a DNA molecule, often but not necessarily the same as the enzyme's recognition site (i.e. a restriction enzyme may recognize one motif, known as a restriction site, and cleave at another).[5]
- TATA box
- A highly conserved non-coding DNA sequence containing a consensus of repeating T and A base pairs that is commonly found in promoter regions of genes in archaea and eukaryotes. The TATA box often serves as the site of initiation of transcription or as a binding site for transcription factors.
- taxis
- A directional response by a cell or a population of cells to a specific stimulus; a movement or other activity occurring in a non-random direction and dependent on the direction from which the stimulus originated.[3] This contrasts with kinesis, a response without directional bias.
- TCA
- See citric acid cycle.
- telestability
- The transmission of three‐dimensional structural stability from a stable part of a macromolecule to a distal part of the same molecule, especially an inherently less stable part.[5] Instability may also be transmitted in this way, e.g. destabilization of the DNA double helix may occur at a locus that is relatively distant from the binding site of a DNA-binding protein.[13]
- telocentric
- (of a linear chromosome or chromosome fragment) Having a centromere positioned at the terminal end of the chromosome (near or within the telomere), resulting in only a single arm.[6] Compare acrocentric.
- telomere
- A region of repetitive nucleotide sequences at each end of a linear chromosome which protects the end of the chromosome from deterioration and from fusion with other chromosomes. Since each round of replication results in the shortening of the chromosome, telomeres act as disposable buffers which are sacrificed to perpetual truncation instead of nearby genes; telomeres can also be lengthened by the enzyme telomerase.
- telomeric silencing
- The repression of transcription of genes in regions adjacent to telomeres. Telomeres also appear to reduce the accessibility of subtelomeric chromatin to modification by DNA methyltransferases.[13]
- telophase
- The final stage of cell division in both mitosis and meiosis, occurring after anaphase and before or simultaneously with cytokinesis, during which a nuclear membrane is synthesized around each set of chromatids, nucleoli are reassembled, and the mitotic spindle is disassembled. Following cytokinesis, the new daughter cells resume interphase.
- template strand
- The strand of a double-stranded DNA molecule which is used as a template for RNA synthesis during transcription. The sequence of the template strand is complementary to the resulting RNA transcript. Contrast coding strand; see also sense.
- terminalization
- In cytology, the progressive shift of chiasmata from their original to more distal positions as meiosis proceeds through diplonema and diakinesis.[13]
- termination codon
- See stop codon.
- terminator
- A DNA sequence or its RNA complement which signals the termination of transcription by triggering processes that ultimately arrest the activity of RNA polymerase or otherwise cause the release of the RNA transcript from the transcriptional complex. Terminator sequences are usually found near the 3'-ends of the coding sequences of genes or operons. They generally function after being themselves transcribed into the nascent RNA strand, whereupon the part of the strand containing the sequence either directly interacts with the transcriptional complex or forms a secondary structure such as a hairpin loop which signals the recruitment of enzymes that promote its disassembly.[13]
- tetramer
- A molecular aggregate consisting of four subunits.[2] The term is often used to refer to protein complexes composed of four proteins, e.g. haemoglobin, or to individual proteins composed of four polypeptides. Compare monomer, dimer, and trimer.
- three-prime end
- See 3'-end.
- three-prime untranslated region
- See 3' untranslated region.
- thymidine (T, dT)
- One of the four standard nucleosides used in DNA molecules, consisting of a thymine base with its N9 nitrogen bonded to the C1 carbon of a deoxyribose sugar. The prefix deoxy- is commonly omitted, since there are no ribonucleoside analogs of thymidine used in RNA, where it is replaced with uridine instead.
- thymine (T)
- A pyrimidine nucleobase used as one of the four standard nucleobases in DNA molecules. Thymine forms a base pair with adenine. In RNA, thymine is not used at all, and is instead replaced with uracil.
- thymine dimer
- See pyrimidine dimer.
- tight junction
- A type of specialized intercellular junction characterized by very close contact between the plasma membranes of adjacent cells, which are held together by large multiprotein complexes that completely or nearly completely occlude the passage of water and solutes through the intercellular space between cells. Tight junctions occur in many vertebrate tissues, especially between the epithelial and endothelial cells that line the surfaces of most organs and vessels. These cells are completely encircled by tight junctions which create a gasket-like seal that separates each cell's plasma membrane into apical and basolateral domains and prevents the exchange of extracellular materials between them.[5][3]
- tissue
- In a multicellular organism, a contiguous aggregation of cells held together by a common extracellular matrix and specialized to perform a particular function. Some tissues are composed primarily of a single cell type; others are a heterogeneous mixture of many cell types.[2] Tissues represent a level of multicellular organization between that of individual cells and that of organs, which may be composed of one or more distinct types of tissue.[3]
- tissue culture
- The growth and maintenance, or "culturing", of multicellular tissues, or of cells harvested from tissues, under carefully controlled conditions in vitro, in the strictest sense by taking a piece of explanted tissue directly from a living plant or animal and maintaining it outside of the body of the source organism. In common usage, the term may also refer to cell culture in general, especially when growing certain cell types which have been harvested from tissues but dispersed from their original tissue-specific organization into a population of more or less independently growing cells.[3]
- tissue-specific gene expression
- Gene function and expression which is restricted to a particular tissue or cell type. Tissue-specific expression is usually the result of an enhancer which is activated only in the proper cell type.
- tonicity
- A measure of the effective osmotic pressure gradient of one solution relative to another solution, used especially to describe the water potential that exists between two aqueous solutions separated by a semipermeable membrane (as with a cell, where the intracellular cytosol is separated from the extracellular fluid by the plasma membrane). Tonicity depends on the relative concentrations of solutes on either side of the membrane, which determine the direction and extent to which solvent molecules move across the membrane by osmosis; it is affected only by those solutes which cannot cross the membrane, as those which can cross freely can achieve equilibrium without any net movement of solute. The extracellular environment is commonly described as hypotonic, hypertonic, or isotonic with respect to the intracellular environment.
- tonoplast
- See vacuole.
- topoisomerase
- Any of a class of DNA-binding enzymes which catalyze changes in the topological state of a double-stranded DNA molecule by nicking or cutting the sugar-phosphate backbone of one or both strands, relaxing the torsional stress inherent in the double helix and unwinding or untangling the paired strands before re-ligating the nicks. This process is usually necessary prior to replication and transcription. Topoisomerases thereby convert DNA between its relaxed and supercoiled, linked and unlinked, and knotted and unknotted forms without changing the sequence or overall chemical composition, such that the substrate and product molecules are structural isomers, differing only in their shape and their twisting, linking, and/or writhing numbers.
- totipotency
- A state of cell potency in which a cell or nucleus fully retains the ability to differentiate into all of the cell types represented in the adult organism, or to give rise to all of these cell types upon transplantation into an appropriate cytoplasm (as in nuclear transfer). Such cells or nuclei are said to be totipotent. The zygote that serves as the progenitor cell for sexually reproducing multicellular organisms is the archetypal totipotent cell; almost all of the cells into which it ultimately differentiates are not totipotent, though some cells such as stem cells remain totipotent or pluripotent throughout the organism's life.[2]
- tracer
- A molecule or a specific atom within a molecule that has been chemically or radioactively labelled so that it can be easily tracked or followed through a biochemical process or located in a cell or tissue.[4]
- trailer sequence
- See 3' untranslated region.
- trans
- On the opposite side; across from; acting from a different molecule. Contrast cis.
- trans-acting
- Affecting a gene or sequence on a different nucleic acid molecule or strand. A locus or sequence within a particular DNA molecule such as a chromosome is said to be trans-acting if it or its products influence or act upon other sequences located relatively far away or on an entirely different molecule or chromosome. For example, a DNA-binding protein acts "in trans" if it binds to or interacts with a sequence located on any strand or molecule different from the one on which it is encoded. Contrast cis-acting.
- trans-splicing
- A form of RNA splicing in which different RNA transcripts, synthesized in separate transcription events, are spliced together into a single, continuous transcript. This contrasts with the more conventional "cis-splicing", where segments of the same transcript are excised or re-arranged.[3]
- transactivation
- An experimental approach to artificially control gene expression by introducing a transactivator gene under the control of an inducible promoter into a genome. The transactivator encodes a transcription factor capable of acting in trans upon one or more other genes by recognizing and specifically binding their promoters; thus by inducing the transactivator gene, the expression of many other genes can be experimentally manipulated.[3]
- transcribed spacer
- A spacer sequence that is transcribed and thus included in the primary ribosomal RNA transcript (as opposed to a non-transcribed spacer) but subsequently excised and discarded during the maturation of functional RNAs of the ribosome.[13]
- transcript
- A product of transcription; that is, any RNA molecule which has been synthesized by RNA polymerase using a complementary DNA molecule as a template. When transcription is completed, transcripts separate from the DNA and become independent primary transcripts. Particularly in eukaryotes, multiple post-transcriptional modifications are usually necessary for raw transcripts to be converted into stable and persistent molecules, which are then described as mature, though not all transcribed RNAs undergo maturation. Many transcripts are accidental, spurious, incomplete, or defective; others are able to perform their functions immediately and without modification, such as certain non-coding RNAs.
- transcript of unknown function (TUF)
- Any RNA transcript whose function is unclear. Such transcripts may include functional non-coding RNAs which have not yet been studied in detail as well as spurious transcripts without any definite function. The DNA sequences from which TUFs are transcribed are generally located in intergenic or intronic regions of the genome. See also junk RNA.
- transcriptase
- See RNA polymerase.
- transcription
- The first step in the process of gene expression, in which an RNA molecule, known as a transcript, is synthesized by enzymes called RNA polymerases using a gene or other DNA sequence as a template. Transcription is a critical and fundamental process in all living organisms and is necessary in order to make use of the information encoded within a genome. All classes of RNA must be transcribed before they can exert their effects upon a cell, though only messenger RNA (mRNA) proceeds to translation to produce a functional protein, whereas the many types of non-coding RNA fulfill their duties without being translated. Transcription is also not always beneficial for a cell: when it occurs at the wrong time or at a functionless locus, or when mobile elements or infectious pathogens utilize the host's transcription machinery, the resulting transcripts (not to mention the waste of valuable energy and resources) are often harmful to the host cell or genome.
- transcription factor (TF)
- Any protein that controls the rate of transcription of genetic information from DNA to RNA by binding to a specific DNA sequence and promoting or blocking the recruitment of RNA polymerase to nearby genes. Transcription factors can effectively turn "on" and "off" specific genes in order to make sure they are expressed at the right times and in the right places; for this reason, they are a fundamental and ubiquitous mechanism of gene regulation.
- transcription start site (TSS)
- The specific location within a gene at which RNA polymerase begins transcription, defined by the specific nucleotide or codon corresponding to the first ribonucleotide(s) to be assembled in the nascent transcript (which is not necessarily the same as the first codon to be translated). This site is usually considered the beginning of the coding sequence and is the reference point for numbering the individual nucleotides within a gene. Nucleotides upstream of the start site are assigned negative numbers and those downstream are assigned positive numbers, which are used to indicate the positions of nearby sequences or structures relative to the TSS. For example, the binding site for RNA polymerase might be a short sequence immediately upstream of the TSS, from approximately -80 to -5, whereas an intron within the coding region might be defined as the sequence starting at nucleotide +207 and ending at nucleotide +793.
- transcription unit
- The segment of DNA between the initiation site and the termination site of transcription, containing the coding sequences for one or more genes. All genes within a transcription unit are transcribed together into a single transcript during a single transcription event; the resulting polycistronic RNA may subsequently be cleaved into separate RNAs, or may be translated as a unit and then cleaved into separate polypeptides.[13]
- transcriptional bursting
- The intermittent nature of transcription and translation mechanisms. Both processes occur in "bursts" or "pulses", with periods of gene activity separated by irregular intervals.
- transcriptome
- The entire set of RNA molecules (often referring to all types of RNA but sometimes exclusively to messenger RNA) that is or can be expressed by a particular genome, cell, population of cells, or species at a particular time or under particular conditions. The transcriptome is distinct from the exome and the translatome.
- transcriptomics
- The study of the transcriptome of a particular genome, cell, or organism, i.e. the sum total of all of the RNA transcripts produced from it by transcription. Transcriptomics technologies allow scientists to isolate and sequence transcriptomes, which can then be mapped to the genome to determine which genes are being expressed or which cellular processes are active and which are dormant at a given time.
- transcytosis
- The transport of molecules across the interior of a cell, i.e. through the cytoplasm, especially a polarized cell such as an epithelial cell with contrasting apical and basal surfaces, thereby providing a spatially oriented transport system. Molecules undergoing transcytosis are usually contained within vesicles.[5]
- transductant
- A cell which has undergone transduction and been successfully transduced.
- transduction
- The transfer of genetic material between cells by a virus or viral vector, either naturally or artificially.
- transfectant
- A cell which has undergone transfection and been successfully transfected.
- transfection
- The deliberate experimental introduction of exogenous nucleic acids into a cell or embryo. In the broadest sense the term may refer to any such transfer and is sometimes used interchangeably with transformation, though some applications restrict the usage of transfection to the introduction of naked or purified non-viral DNA or RNA into cultured eukaryotic cells (especially animal cells) resulting in the subsequent incorporation of the foreign DNA into the host genome or the non-hereditary modification of gene expression by the foreign RNA. As a contrast to both standard non-viral transformation and transduction, transfection has also occasionally been used to refer to the uptake of purified viral nucleic acids by bacteria or plant cells without the aid of a viral vector.[13]
- transfer RNA (tRNA)
- A special class of RNA molecule, typically 76 to 90 nucleotides in length, that serves as a physical adapter allowing mRNA transcripts to be translated into sequences of amino acids during protein synthesis. Each tRNA contains a specific anticodon triplet corresponding to an amino acid that is covalently attached to the tRNA's opposite end; as translation proceeds, tRNAs are recruited to the ribosome, where each mRNA codon is paired with a tRNA containing the complementary anticodon. Depending on the organism, cells may employ as many as 41 distinct tRNAs with unique anticodons; because of codon degeneracy within the genetic code, several tRNAs containing different anticodons carry the same amino acid.
- transferase
- Any of a class of enzymes which catalyze the chemical transfer of a functional group or substituent from one molecule to another.[2] For example, acetyltransferases catalyze the movement of an acetyl group in a process known as acetylation; methyltransferases catalyze the movement of one or more methyl groups in a process known as methylation.
- transfer-messenger RNA (tmRNA)
- A type of RNA molecule in some bacteria which has dual tRNA-like and mRNA-like properties, allowing it to simultaneously perform a number of different functions during translation.
- transformant
- A cell or organism which has taken up extracellular DNA by transformation and which can express genes encoded by it.
- transformation
- transgene
- Any gene or other segment of genetic material that has been isolated from one organism and then transferred either naturally or by any of a variety of genetic engineering techniques into another organism, especially one of a different species. Transgenes are usually introduced into the second organism's germ line. They are commonly used to study gene function or to confer an advantage not otherwise available in the unaltered organism.
- transition
- A point mutation in which a purine nucleotide is substituted for another purine (A ↔ G) or a pyrimidine nucleotide is substituted for another pyrimidine (C ↔ T). Contrast transversion.
- translation
- The second step in the process of gene expression, in which the messenger RNA transcript produced during transcription is read by a ribosome to produce a functional protein.
- translatome
- The entire set of messenger RNA molecules that are translated by a particular genome, cell, tissue, or species at a particular time or under particular conditions. Like the transcriptome, it is often used as a proxy for quantifying levels of gene expression, though the transcriptome also includes many RNA molecules that are never translated.
- translocation
- A type of chromosomal abnormality caused by the structural rearrangement of large sections of one or more chromosomes. There are two main types: reciprocal and Robertsonian.
- transmembrane protein
- See integral polytopic protein.
- transmission genetics
- The branch of genetics that studies the mechanisms involved in the transfer of genes from parents to offspring.[13]
- transport protein
- Any transmembrane protein which functions by permitting the movement of particular molecules, proteins, or other substances across a membrane, either actively or passively and in either or both directions (by which they may be further subclassified into uniporters, antiporters, and symporters).[3] Channel proteins and nuclear pores are examples of transport proteins.
- transporter
- See transport protein.
- transposable element (TE)
- Any of a diverse variety of selfish mobile genetic elements consisting of self-acting DNA sequences capable of replicating themselves semi-autonomously and inserting into random or specific sites within a host genome, a process known as transposition. Transposons contain one or more genes which encode enzymes known as transposases capable of recognizing sequences within a flanking pair of inverted repeats, such that the enzymes effectively catalyze their own replication, excision, and/or re-insertion into other DNA molecules by any of various mechanisms.[3]
- transposase
- Any of a class of self-acting enzymes capable of binding to the flanking sequences of the transposable element which encodes them and catalyzing its movement to another part of the genome, typically by an excision/insertion mechanism or a replicative mechanism, in a process known as transposition.
- transposition
- The process by which a nucleic acid sequence known as a transposable element changes its position within a genome, either by excising and re-inserting itself at a different locus (cut-and-paste) or by duplicating itself and inserting into another locus without moving the original element from its original locus (copy-paste). These reactions are catalyzed by an enzyme known as a transposase which is encoded by a gene within the transposable element itself; thus the element's products are self-acting and can autonomously direct their own replication. Transposed sequences may re-insert at random loci or at sequence-specific targets, either on the same DNA molecule or on different molecules.
- transvection
- transversion
- A point mutation in which a purine nucleotide is substituted for a pyrimidine nucleotide, or vice versa (e.g. A ↔ C or A ↔ T). Contrast transition.
- tricarboxylic acid cycle (TCA)
- See citric acid cycle.
- triglyceride
- Any of a class of chemical compounds which are ester derivatives of glycerol, consisting of a glycerol backbone connected to any three fatty acid substituents via ester bonds. Triglycerides are one of three major classes of esters formed by fatty acids in biological systems, along with phospholipids and cholesteryl esters. They are the primary constituent of adipose tissue in vertebrates.
- trimer
- A molecular aggregate consisting of three subunits.[31] The term is often used to refer to protein complexes composed of three proteins, e.g. many membrane porins, or to individual proteins composed of three polypeptides. Compare monomer, dimer, and tetramer.
- trinucleotide repeat
- Any sequence in which an individual nucleotide triplet is repeated many times in tandem, whether in a gene or non-coding sequence. At most loci some degree of repetition is normal and harmless, but mutations which cause specific triplets (especially those of the form CnG) to increase in copy number above the normal range are highly unstable and responsible for a variety of genetic disorders.
- triplet
- A unit of three successive nucleotides in a DNA or RNA molecule.[13] A triplet within a coding sequence that codes for a specific amino acid is known as a codon.
- trisomy
- A type of polysomy in which a diploid cell or organism has three copies of a particular chromosome instead of the normal two.
- tRNA
- See transfer RNA.
- tRNA-ligase
- See aminoacyl-tRNA synthetase.
- tropism
- The directional growth or movement of a cell or organism in response to a stimulus, e.g. light, heat, the pull of gravity, or the presence of a particular chemical, such that the response is dependent on the direction of the stimulus (as opposed to a non-directional nastic response). Positive tropism is growth or movement toward the stimulus; negative tropism is away from the stimulus.[2] See also taxis and kinesis.
- tumorigenesis
- turgor pressure
- The force within a cell which pushes the plasma membrane against the cell wall,[32] a type of hydrostatic pressure influenced by the osmotic flow of water into and out of the cell. Turgidity is observed in plants, fungi, bacteria, and some protists with cell walls, but generally not in animal cells.
- turnover number
- In enzymology, a measure of the rate at which a particular enzyme catalyzes a particular biochemical reaction, usually expressed as the average number of substrate molecules it is capable of converting into reaction products per unit time at a given concentration of enzyme.[33]
- twisting number


U
[edit]- ubiquitin (Ub)
- A small protein of 76 amino acids found in great quantities (ubiquitously) in all eukaryotic cells, employed chiefly as a post-translational protein tag, by which its C-terminal glycine residue is covalently bonded to electrically charged residues within other proteins or polypeptides, a process known as ubiquitination. Ubiquitin tags have functions in the heat-shock response, protein sorting, proteolysis, membrane trafficking, cell signaling, regulation of the cell cycle, X chromosome inactivation, and histone modification, among others.[3]
- ubiquitination
- The labelling of a biomolecule (often another protein) by covalently attaching a ubiquitin protein to it—generally via the formation of an amide bond between the ubiquitin's C-terminal glycine and positively charged side chains (often lysine or arginine residues) of the labelled molecule, an ATP-dependent reaction catalyzed by ubiquitin-conjugating enzymes[3]—thus making it identifiable to molecules capable of recognizing ubiquitin epitopes. Ubiquitination is a widely used post-translational modification by which proteins are tagged; the attachment of a single ubiquitin molecule (monoubiquitination) can variously activate or inhibit a protein's activity, while the attachment of a chain of multiple consecutively linked ubiquitin molecules (polyubiquitination) commonly targets the protein for degradation by proteasomes.
- umber
- See opal.
- uncharged tRNA
- A transfer RNA without an attached amino acid. Contrast charged tRNA.
- underwinding
- See negative supercoiling.
- unequal crossing over
- uniporter
- A type of transport protein which catalyzes the movement of a single, specific solute or chemical species across a lipid membrane in either direction.[4] Contrast antiporter and symporter.
- unique DNA
- A class of DNA sequences determined by C0t analysis to be present only once in the analyzed genome, as opposed to repetitive sequences. Most structural genes and their introns are unique.[13]
- unstable mutation
- A mutation with a high frequency of reversion.[13]
- untranslated region (UTR)
- Any non-coding sequence which is transcribed along with a protein-coding sequence, and thus included within a messenger RNA, but which is not ultimately translated during protein synthesis. A typical mRNA transcript includes two such regions: one immediately upstream of the coding sequence, known as the 5' untranslated region (5'-UTR), and one downstream of the coding sequence, known as the 3' untranslated region (3'-UTR). These regions are not removed during post-transcriptional processing (unlike introns) and are usually considered distinct from the 5' cap and the 3' polyadenylated tail (both of which are later additions to a primary transcript and not themselves products of transcription). UTRs are a consequence of the fact that transcription usually begins considerably upstream of the start codon of the coding sequence and terminates long after the stop codon has been transcribed, whereas translation is more precise. They often include motifs with regulatory functions.
- upregulation
- Any process, natural or artificial, which increases the level of gene expression of a certain gene. A gene which is observed to be expressed at relatively high levels (such as by detecting higher levels of its mRNA transcripts) in one sample compared to another sample is said to be upregulated. Contrast downregulation.
- upstream
- Towards or closer to the 5'-end of a chain of nucleotides, or the N-terminus of a peptide chain. Contrast downstream.
- upstream activating sequence (UAS)
- A type of cis-acting regulatory element found in the DNA of yeast such as Saccharomyces cerevisiae, usually a few hundred base pairs upstream of the transcription initiation site within the promoter of a protein-coding gene, which helps to increase the gene's expression by serving as a binding site for transcriptional transactivators, analogous to the function of an enhancer in multicellular eukaryotes.[34][5]
- uracil (U)
- A pyrimidine nucleobase used as one of the four standard nucleobases in RNA molecules. Uracil forms a base pair with adenine. In DNA, uracil is not used at all, and is instead replaced with thymine.
- uridine (U, Urd)
- One of the four standard nucleosides used in RNA molecules, consisting of a uracil base with its N9 nitrogen bonded to the C1 carbon of a ribose sugar. In DNA, uridine is replaced with thymidine.

V
[edit]- vacuole
- Any of a class of enclosed, fluid-filled compartments present in many eukaryotic cells as well as bacteria, often large and conspicuous under the microscope and serving any of a huge variety of functions, including acting as a resizable reservoir for the storage of water, metabolic waste, toxins, or foreign material; maintaining cellular homeostasis and hydrostatic pressure; supporting immune functions; housing symbiotic bacteria; and assisting in the degradation and recycling of old cellular components.[2]
- variable number tandem repeat (VNTR)
- Any of a class of tandem repeats for which the copy number of the repeated sequence at a particular locus tends to vary between individuals of the same species. VNTRs may occur throughout the genome, both within and outside of coding DNA, and if the copy number is stably inherited may be used in DNA fingerprinting to uniquely identify individuals or to determine their genealogical relatedness to other individuals.
- variegation
- Variation or irregularity in a particular phenotype, especially a conspicuous visible trait such as color or pigmentation, occurring simultaneously in different parts of the same individual organism due to any of a variety of causes, such as X-inactivation, mitotic recombination, transposable element activity, position effects, or infection by pathogens.
- vector
- Any DNA molecule used as a vehicle to artificially transport foreign genetic material into another cell, where it can be replicated and/or expressed. Vectors are typically engineered recombinant DNA sequences consisting of an insert (often a transgene) and a longer "backbone" sequence containing an origin of replication, a multiple cloning site, and a selectable marker. Vectors are widely used in molecular biology laboratories to isolate, clone, or express the insert in the target cell.
- vectorization
- vegetal cell
- See somatic cell.
- vesicle
- Any membrane-bound space completely enclosed by its own membrane, which is separate though usually derived from other membranes (often the cell membrane) either by budding or by mechanical disruption such as sonication.[3] The term is applied to many different structures but especially to the small, roughly spherical compartments created during endocytosis and exocytosis, as well as to lysosomes and various other small intracellular or extracellular organelles.[2]
W
[edit]- Warburg effect
- western blotting
- A blotting method used for detecting and identifying specific proteins in heterogeneous biological samples. The technique involves separating proteins by size with gel electrophoresis and then immobilizing them upon a nitrocellulose, nylon, or other synthetic membrane, after which they may be visualized by autoradiography or by labelling with chemiluminescent, radioactive, or enzyme-linked antibodies, lectins, or other specific binding agents.[5] Compare Southern blotting, northern blotting, and eastern blotting.
- whole genome sequencing (WGS)
- The process of determining the entirety or near-entirety of the DNA sequences comprising an organism's genome with a single procedure or experiment, generally inclusive of all chromosomal and extrachromosomal (e.g. mitochondrial) DNA.
- wild type (WT)
- The phenotype of the typical form of a species as it occurs in nature; a product of the standard "normal" allele at a given locus, as opposed to that produced by a non-standard mutant allele.
- wobble base pairing
- writhing number (W)
- An index of the superhelical coiling of a DNA molecule. The writhing number does not have a precise quantitative definition but instead represents the degree of supercoiling. Together, the writhing number and the twisting number determine the linking number.[5]

X
[edit]- X chromosome
- One of two sex chromosomes present in organisms which use the XY sex-determination system, and the only sex chromosome in the X0 system. The X chromosome is found in both males and females and typically contains much more gene content than its counterpart, the Y chromosome.
- X-inactivation
- The process by which one of the two copies of the X chromosome is silenced by being irreversibly condensed into transcriptionally inactive heterochromatin in the cells of female therian mammals. A form of dosage compensation, X-inactivation prevents females from producing twice as many gene products from genes on the X chromosome as males, who only have one copy of the X chromosome. Which X chromosome is inactivated is randomly determined in the early embryo, making it possible for cell lineages with different inactive Xs to exist in the same organism.
- X-linked trait
- A phenotypic trait whose expression is governed or influenced by one or more genes located on the X chromosome (making it a sex-linked trait).
Y
[edit]- Y chromosome
- One of two sex chromosomes present in organisms which use the XY sex-determination system. The Y chromosome is found only in males and is typically much smaller than its counterpart, the X chromosome.
- Y fork
- See replication fork.
- yeast artificial chromosome (YAC)
Z
[edit]- Z-DNA
- zinc finger (ZF)
- A supersecondary polypeptide structural motif and DNA-binding domain occurring in many DNA-binding proteins, characterized by a series of non-adjacent amino acid residues which fold into a three-dimensional arrangement capable of coordinating one or more zinc ions (Zn2+
) between them, thus stabilizing the fold into a definite structure that can interact specifically with other biomolecules such as nucleic acids or other polypeptides. There are many distinct classes of zinc fingers using different ligands and spatial arrangements to achieve coordination; in perhaps the most common variant, a short alpha helix is oriented antiparallel to a β sheet, with two histidine residues in the former and two cysteines in the latter forming the coordination complex. Zinc fingers bind DNA as the primary functional domain of many transcription factors. - zonula adherens
- See adherens junction.
- zonula occludens
- See tight junction.
- zygonema
- In meiosis, the second of five substages of prophase I, following leptonema and preceding pachynema. During zygonema, synapsis occurs, physically binding homologous chromosomes to each other, and the cell's centrosome divides into two daughter centrosomes, each containing a single centriole.[13]
- zygosity
- The degree to which multiple copies of a gene, chromosome, or genome have the same genetic sequence; e.g. in a diploid organism with two complete copies of its genome (one maternal and one paternal), the degree of similarity of the alleles present in each copy. Individuals carrying two different alleles for a particular gene are said to be heterozygous for that gene; individuals carrying two identical alleles are said to be homozygous for that gene. Zygosity may also be considered collectively for a group of genes, or for the entire set of genes and genetic loci comprising the genome.
- zygote
- A type of eukaryotic cell formed as the direct result of a fertilization event between two gametes. In multicellular organisms, the zygote is the earliest developmental stage.
See also
[edit]References
[edit]- ^ "Talking Glossary of Genomic and Genetic Terms". genome.gov. 8 October 2017. Retrieved 8 October 2017.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab MacLean, Norman (1987). Dictionary of Genetics & Cell Biology. New York: New York University Press. ISBN 0-8147-5438-4.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac ad ae af ag ah ai aj ak al am an ao ap Lackie, J. M. (2013). The Dictionary of Cell and Molecular Biology (5th ed.). Amsterdam: Academic Press/Elsevier. ISBN 978-0-12-384931-1.
- ^ a b c d e f g h i j k l m n o p Alberts, Bruce; Johnson, Alexander; Lewis, Julian; Raff, Martin; Roberts, Keith; Walter, Peter (2002). "Glossary". Molecular Biology of the Cell (Available from the National Center for Biotechnology Information) (4th ed.). New York: Garland Science.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac ad ae af ag ah ai aj ak al am an ao ap aq ar Cammack, Richard; Atwood, Teresa; Campbell, Peter; Parish, Howard; Smith, Anthony; Vella, Frank; Stirling, John, eds. (2008). Oxford Dictionary of Biochemistry and Molecular Biology (2nd ed.). Oxford: Oxford University Press. ISBN 978-0-19-172764-1.
- ^ a b c Klug, William S.; Cummings, Michael R. (1986). Concepts of Genetics (2nd ed.). Glenview, Ill.: Scott, Foresman and Company. ISBN 0-673-18680-6.
- ^ Rayner TF; Rocca-Serra P; Spellman PT; Causton HC; et al. (2006). "A simple spreadsheet-based, MIAME-supportive format for microarray data: MAGE-TAB". BMC Bioinformatics. 7 489. doi:10.1186/1471-2105-7-489. PMC 1687205. PMID 17087822.
- ^ Yáñez-Mó M, Siljander PR, Andreu Z, Zavec AB, Borràs FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, Colás E, Cordeiro-da Silva A, Fais S, Falcon-Perez JM, Ghobrial IM, Giebel B, Gimona M, Graner M, Gursel I, Gursel M, Heegaard NH, Hendrix A, Kierulf P, Kokubun K, Kosanovic M, Kralj-Iglic V, Krämer-Albers EM, Laitinen S, Lässer C, Lener T, Ligeti E, Linē A, Lipps G, Llorente A, Lötvall J, Manček-Keber M, Marcilla A, Mittelbrunn M, Nazarenko I, Nolte-'t Hoen EN, Nyman TA, O'Driscoll L, Olivan M, Oliveira C, Pállinger É, Del Portillo HA, Reventós J, Rigau M, Rohde E, Sammar M, Sánchez-Madrid F, Santarém N, Schallmoser K, Ostenfeld MS, Stoorvogel W, Stukelj R, Van der Grein SG, Vasconcelos MH, Wauben MH, De Wever O (2015). "Biological properties of extracellular vesicles and their physiological functions". J Extracell Vesicles. 4 27066. doi:10.3402/jev.v4.27066. PMC 4433489. PMID 25979354.
- ^ a b c d e f g h i j k Rieger, Rigomar (1991). Glossary of Genetics: Classical and Molecular (5th ed.). Berlin: Springer-Verlag. ISBN 3-540-52054-6.
- ^ Functional Genomics Data Society (June 2012). "Minimum Information about a high-throughput SEQuencing Experiment". Archived from the original on 2022-12-06. Retrieved 2022-11-13.
- ^ Oliver S (2003). "On the MIAME Standards and Central Repositories of Microarray Data". Comparative and Functional Genomics. 4 (1): 1. doi:10.1002/cfg.238. PMC 2447402. PMID 18629115.
- ^ Brazma A (2009). "Minimum Information About a Microarray Experiment (MIAME)--successes, failures, challenges". ScientificWorldJournal. 9: 420–3. doi:10.1100/tsw.2009.57. PMC 5823224. PMID 19484163.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac ad ae af ag ah ai aj ak al am an ao ap King, Robert C.; Stansfield, William D.; Mulligan, Pamela K. (2006). A Dictionary of Genetics (7th ed.). Oxford: Oxford University Press. ISBN 978-0-19-530762-7.
- ^ Astbury, W. T. (June 1961). "Molecular Biology or Ultrastructural Biology ?". Nature. 190 (4781): 1124. Bibcode:1961Natur.190.1124A. doi:10.1038/1901124a0. ISSN 1476-4687. PMID 13684868. S2CID 4172248.
- ^ Klermund, Julia; Rhiel, Manuel; Kocher, Thomas; Chmielewski, Kay Ole; Bischof, Johannes; Andrieux, Geoffroy; el Gaz, Melina; Hainzl, Stefan; Boerries, Melanie; Cornu, Tatjana I.; Koller, Ulrich; Cathomen, Toni (May 2024). "On- and off-target effects of paired CRISPR-Cas nickase in primary human cells". Molecular Therapy. 32 (5): 1298–1310. doi:10.1016/j.ymthe.2024.03.006. PMC 11081867. PMID 38459694.
- ^ "7.6D: The Incorporation of Nonstandard Amino Acids". LibreTexts Biology.
- ^ Hartwell, Leland; Hood, L.; Goldberg, M.; Reynolds, A.; Silver, L.; Veres, R. (2004). Genetics: From Genes to Genomes. McGraw-Hill. p. 267. ISBN 978-0-07-246248-7. OCLC 50417228.
- ^ a b Lewin, Benjamin (2003). Genes VIII. Upper Saddle River, NJ: Pearson Prentice Hall. ISBN 0-13-143981-2.
- ^ "overexpression". Oxford Living Dictionary. Oxford University Press. 2017. Archived from the original on February 10, 2018. Retrieved 18 May 2017.
The production of abnormally large amounts of a substance which is coded for by a particular gene or group of genes; the appearance in the phenotype to an abnormally high degree of a character or effect attributed to a particular gene.
- ^ "overexpress". NCI Dictionary of Cancer Terms. National Cancer Institute at the National Institutes of Health. 2011-02-02. Retrieved 18 May 2017.
overexpress
In biology, to make too many copies of a protein or other substance. Overexpression of certain proteins or other substances may play a role in cancer development. - ^ Lefranc MP (2013). "Paratope". In Dubitzky W, Wolkenhauer O, Cho KH, Yokota H (eds.). Encyclopedia of Systems Biology. New York, NY: Springer. pp. 1632–1633. doi:10.1007/978-1-4419-9863-7_673. ISBN 978-1-4419-9863-7.
- ^ Punt J, Stranford SA, Jones PP, Owen JA (2019). Kuby immunology (Eighth ed.). New York. ISBN 978-1-4641-8978-4. OCLC 1002672752.
{{cite book}}: CS1 maint: location missing publisher (link) - ^ Lagunas R (1981-01-01). "Is Saccharomyces cerevisiae a typical facultative anaerobe?". Trends in Biochemical Sciences. 6: 201–203. doi:10.1016/0968-0004(81)90073-6. ISSN 0968-0004.
- ^ Goñi FM (2002). "Non-permanent proteins in membranes: when proteins come as visitors (Review)". Molecular Membrane Biology. 19 (4): 237–245. doi:10.1080/0968768021000035078. PMID 12512770. S2CID 20892603.
- ^ Johnson JE, Cornell RB (1999). "Amphitropic proteins: regulation by reversible membrane interactions (review)". Molecular Membrane Biology. 16 (3): 217–235. doi:10.1080/096876899294544. PMID 10503244.
- ^ SwissBioPics. "Eukaryota cell". www.swissbiopics.org. Swiss Institute of Bioinformatics (SIB).
- ^ Tambe, D.T., Hardin, C., Angelini, T.E., Rajendran, K., Park, C.Y., Serra-Picamal, X., Zhou, E.H., Zaman, M.H., Butler, J.P., Weitz, D.A., Fredberg JJ, Trepat, X, Collective cell guidance by cooperative intercellular forces. Nature Materials,10(6), 469–475, 2011, doi:10.1038/nmat3025.
- ^ Lodish, Berk, Kaiser, Krieger, Bretscher, Ploegh, Martin, Yaffe, Amon (2021). Molecular Cell Biology (9th ed.). New York, NY: W.H. Freeman and Company. ISBN 978-1-319-20852-3.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ "Nomenclature of glycoproteins, glycopeptides and peptidoglycans, Recommendations 1985". www.qmul.ac.uk. Retrieved 16 March 2021.
- ^ Will CL, Lührmann R (July 2011). "Spliceosome structure and function". Cold Spring Harbor Perspectives in Biology. 3 (7) a003707. Bibcode:2011CSHPB...303707W. doi:10.1101/cshperspect.a003707. PMC 3119917. PMID 21441581.
- ^ "Levels of Protein Organization: A 2014 Foundations of Medicine eLAB". comis.med.uvm.edu. University of Vermont, Larner College of Medicine.
- ^ Pritchard, Jeremy (2001). "Turgor Pressure". Encyclopedia of Life Sciences. American Cancer Society. doi:10.1038/npg.els.0001687. ISBN 978-0-470-01590-2.
- ^ Smejkal, Gary B.; Kakumanu, Srikanth (3 July 2019). "Enzymes and their turnover numbers". Expert Review of Proteomics. 16 (7): 543–544. doi:10.1080/14789450.2019.1630275. PMID 31220960.
- ^ Webster, Nocholas; Jin, Jia Rui; Green, Stephen; Hollis, Melvyn; Chambon, Pierre (29 January 1988). "The Yeast UASG is a transcriptional enhancer in human hela cells in the presence of the GAL4 trans-activator". Cell. 52 (2): 169–178. doi:10.1016/0092-8674(88)90505-3. PMID 2830022. S2CID 26819676.
Further reading
[edit]- Budd, A. (2012). "Introduction to genome biology: features, processes, and structures". Evolutionary Genomics. Methods in Molecular Biology. Vol. 855. pp. 3–4. doi:10.1007/978-1-61779-582-4_1. ISBN 978-1-61779-581-7. PMID 22407704.
External links
[edit]Glossary of cellular and molecular biology (M–Z)
View on Grokipediafrom Grokipedia
M
Membrane and Organelle Structures
In cellular and molecular biology, membrane and organelle structures form the foundational compartments that enable eukaryotic cells to organize biochemical processes, maintain homeostasis, and respond to environmental cues. These structures, primarily composed of macromolecules and lipid bilayers, compartmentalize the cytoplasm into specialized regions, facilitating selective transport, energy production, and degradation pathways. Key components include lipid-based barriers embedded with proteins, as well as membrane-enclosed organelles that house enzymatic reactions essential for cellular function. Macromolecules are large, complex molecules typically formed by the polymerization of smaller monomeric units, such as amino acids into proteins, nucleotides into nucleic acids, and monosaccharides into polysaccharides, playing critical roles in cellular architecture, catalysis, and information storage.[6] These polymers, often exceeding thousands of daltons in molecular weight, provide structural integrity to membranes and organelles while enabling dynamic interactions like folding and assembly into functional complexes.[7] In the context of membrane and organelle structures, macromolecules such as proteins and lipids integrate to form stable yet fluid barriers that support compartmentalization.[8] Membrane refers to the plasma membrane or intracellular membranes that enclose cells and organelles, consisting of a phospholipid bilayer that acts as a selective permeability barrier, regulating the influx and efflux of ions, nutrients, and waste products.[9] This structure separates the cell's interior from the extracellular environment while allowing communication through embedded proteins, thereby maintaining cellular integrity and enabling processes like endocytosis and exocytosis.[10] Membranes are dynamic, with fluidity influenced by temperature and lipid composition, which is essential for organelle biogenesis and vesicle trafficking.[11] Membrane lipid denotes the amphipathic molecules, primarily phospholipids and sterols like cholesterol, that self-assemble into the hydrophobic core of the lipid bilayer, providing a semi-permeable matrix that excludes water-soluble substances while allowing lateral diffusion of embedded components.[12] These lipids, with hydrophilic heads and hydrophobic tails, spontaneously form bilayers in aqueous environments, contributing approximately 50% of the membrane's mass and modulating membrane thickness and curvature for organelle shaping.[13] Cholesterol, in particular, intercalates between phospholipids to reduce fluidity at physiological temperatures, enhancing membrane stability in structures like the plasma membrane and mitochondrial inner membrane.[14] Membrane protein encompasses integral proteins, which span the lipid bilayer via transmembrane domains, and peripheral proteins, which associate loosely with the membrane surface, collectively facilitating transport, enzymatic activity, signaling, and structural support within cellular compartments.[15] Integral membrane proteins, such as channels and receptors, often traverse the bilayer multiple times with alpha-helical segments, enabling selective ion permeation and signal transduction across organelles like the endoplasmic reticulum and mitochondria.[16] Peripheral proteins, bound via electrostatic interactions, aid in cytoskeletal anchoring and vesicle docking, contributing to the asymmetry and functionality of membrane-bound structures.[17] Membrane-bound organelle describes intracellular compartments enclosed by one or more lipid bilayers, such as the nucleus, endoplasmic reticulum, and Golgi apparatus, which segregate specialized enzymatic reactions and prevent cross-talk between incompatible biochemical pathways.[18] These organelles create distinct microenvironments by acting as diffusion barriers, with their membranes enriched in specific lipids and proteins that support functions like protein synthesis and lipid modification.[19] In eukaryotic cells, this compartmentalization enhances efficiency, as seen in the endoplasmic reticulum's role in folding secretory proteins away from cytosolic processes.[20] Microbody is a single-membrane-bound organelle, typically 0.5–1.5 μm in diameter, including peroxisomes and glyoxysomes, that houses oxidative enzymes for detoxifying reactive oxygen species and metabolizing fatty acids or photorespiratory intermediates.[21] These organelles contain a dense proteinaceous matrix with catalases and oxidases, enabling the breakdown of hydrogen peroxide generated during beta-oxidation, thus protecting cellular components from oxidative damage.[22] In plant cells, glyoxysomes within microbodies facilitate the conversion of stored lipids to carbohydrates during seed germination, highlighting their role in metabolic compartmentalization.[23] Micronucleus, in protozoans like ciliates (e.g., Paramecium), is a smaller, diploid nucleus distinct from the larger macronucleus, serving as the germline genome for genetic recombination and transmission during conjugation and sexual reproduction.[24] Unlike the transcriptionally active macronucleus, the micronucleus remains dormant during vegetative growth but undergoes precise DNA replication and segregation during cell division, ensuring genetic stability.[25] This dimorphic nuclear organization allows ciliates to separate somatic and reproductive functions within a single cell, a unique adaptation in membrane-bound nuclear structures. Midbody is a transient, microtubule-based structure formed at the intercellular bridge during the final stages of cytokinesis, consisting of bundled central spindle fibers and associated proteins that orchestrate the abscission of daughter cells.[26] Composed of compacted antiparallel microtubules enveloped by a plasma membrane, the midbody recruits endosomal sorting complex required for transport (ESCRT) machinery to constrict and sever the bridge, ensuring complete cell separation.[27] Post-abscission, midbody remnants can persist as signaling organelles, influencing polarity and proliferation in the daughter cells.[28] Mitochondrion is a double-membrane-bound organelle, often termed the powerhouse of the cell, that generates ATP through oxidative phosphorylation via the electron transport chain embedded in its inner membrane cristae.[29] The outer membrane permits free diffusion of small molecules, while the inner membrane, highly folded to increase surface area, maintains a proton gradient essential for ATP synthase activity, coupling it to the tricarboxylic acid cycle in the matrix.[30] Mitochondria also contain their own circular DNA and ribosomes, supporting semi-autonomous replication and protein synthesis for respiratory complexes.[31] Mitophagy is the selective autophagic process that targets dysfunctional or excess mitochondria for lysosomal degradation, involving ubiquitination and engulfment by autophagosomes to maintain mitochondrial quality and prevent oxidative stress accumulation.[32] Triggered by stressors like hypoxia or damaged respiratory chains, mitophagy employs receptors such as BNIP3 or Parkin to tag mitochondria for clearance, thereby preserving cellular energy homeostasis and averting apoptosis.[33] This pathway is crucial for organelle turnover, with defects linked to neurodegenerative diseases due to impaired mitochondrial dynamics.[34]Metabolism and Biochemical Processes
MetabolismMetabolism refers to the entirety of chemical reactions occurring within cells to maintain life, providing energy and enabling the synthesis of cellular components.[35] It is divided into two main categories: anabolism, which consumes energy to construct complex molecules from simpler precursors, such as the assembly of proteins from amino acids; and catabolism, which breaks down macromolecules to release energy, often in the form of ATP, as seen in the degradation of glucose.[36] These interconnected processes ensure cellular homeostasis and respond to environmental cues, with catabolic pathways typically predominating during energy demand to fuel anabolic reactions.[37] Metabolic pathway
A metabolic pathway consists of a sequence of enzyme-catalyzed reactions that transform substrates into products, facilitating the flow of metabolites through interconnected steps.[38] For example, glycolysis is an anaerobic pathway that converts glucose into pyruvate, generating ATP and NADH without oxygen, while the Krebs cycle (also known as the citric acid cycle) oxidizes acetyl-CoA in mitochondria to produce electron carriers for oxidative phosphorylation.[39] These pathways are regulated by enzymes and allosteric effectors to balance energy production and biosynthetic needs, with disruptions often leading to metabolic disorders.[40] Metabolic waste
Metabolic waste encompasses the byproducts of cellular metabolism that accumulate during energy conversion and must be eliminated to avoid toxicity, such as carbon dioxide from oxidative respiration and urea from amino acid catabolism.[41] In vertebrates, urea is synthesized in the liver through the urea cycle, a series of reactions that detoxifies ammonia—a harmful byproduct of protein breakdown—before excretion by the kidneys.[36] These wastes reflect the efficiency of catabolic processes, with their removal essential for maintaining pH balance and preventing cellular stress.[35] Metabolite
A metabolite is a small, low-molecular-weight organic or inorganic molecule that serves as an intermediate, reactant, or product in metabolic pathways, not directly encoded by the genome but generated through enzymatic activity.[42] Examples include glucose-6-phosphate in glycolysis or acetyl-CoA in the Krebs cycle, which link catabolic and anabolic reactions to support energy transfer and biosynthesis.[39] Metabolites dynamically reflect cellular state, influencing signaling and regulation, as their concentrations modulate enzyme kinetics in pathways.[43] Metabolome
The metabolome represents the complete set of all small-molecule metabolites present in a cell, tissue, or organism at a specific time, providing a snapshot of metabolic activity and environmental interactions.[44] Unlike the static genome, the metabolome is highly dynamic, encompassing thousands of compounds like amino acids, lipids, and sugars that vary with physiological conditions.[45] It serves as the downstream readout of genomic and proteomic functions, enabling the study of how perturbations in pathways affect overall cellular health.[46] Methylation
Methylation involves the covalent addition of a methyl group (CH₃) to biomolecules such as DNA, proteins, or metabolites, serving as a key regulatory modification in cellular processes.[47] In DNA, methylation typically occurs at the 5-position of cytosine residues in CpG dinucleotides, recruiting repressive proteins to silence gene expression and maintain epigenetic memory.[48] For metabolites, methylation alters solubility and activity, as in the conversion of norepinephrine to epinephrine, while protein methylation on histones or non-histone targets influences chromatin structure and signaling.[49] This process is reversible and tightly controlled, impacting development, disease, and metabolic adaptation.[50] Methyltransferase
Methyltransferases are a class of transferase enzymes that catalyze the transfer of a methyl group from S-adenosylmethionine (SAM) to acceptor substrates, including nucleic acids, proteins, and small molecules.[51] DNA methyltransferases (DNMTs), such as DNMT1 for maintenance and DNMT3 for de novo methylation, establish epigenetic patterns by modifying cytosines, essential for gene regulation.[52] In metabolic contexts, enzymes like catechol O-methyltransferase (COMT) detoxify catecholamines, highlighting their role in both regulatory and catabolic functions.[53] These enzymes ensure precise control over methylation, with dysregulation linked to cancers and neurological disorders.[54] Mistranslation
Mistranslation refers to errors in protein synthesis where the ribosome incorporates incorrect amino acids into the polypeptide chain, deviating from the mRNA codon sequence.[55] Such errors arise from tRNA misacylation, near-cognate tRNA selection, or ribosomal frameshifting, occurring at rates of about 10⁻⁴ per codon in eukaryotes but increasing under stress.[56] While generally deleterious, leading to misfolded proteins and cellular toxicity, mistranslation can confer adaptive benefits, such as phenotypic variability during environmental challenges.[57] Quality control mechanisms, including editing by aminoacyl-tRNA synthetases and degradation by the proteasome, mitigate these errors to preserve proteome integrity.[58] Monosaccharide
A monosaccharide is the simplest form of carbohydrate, consisting of a single sugar unit with the general formula Cₙ(H₂O)ₙ, serving as a fundamental building block for more complex carbohydrates and a primary energy source.[59] Common examples include glucose (an aldohexose), fructose (a ketohexose), and ribose (a pentose), which exist in linear or cyclic forms and participate in pathways like glycolysis for ATP production.[60] Monosaccharides also contribute to structural roles, such as in nucleotide synthesis, and their phosphorylation facilitates metabolic regulation and transport across membranes.[61] Motor protein
Motor proteins are ATP-dependent enzymes that generate mechanical force for intracellular transport and motility by undergoing conformational changes along cytoskeletal filaments.[62] Key families include kinesins and dyneins, which move cargos along microtubules toward the plus or minus ends, respectively, and myosins, which interact with actin filaments for processes like vesicle trafficking and muscle contraction.[63] These proteins hydrolyze ATP to produce directed movement at speeds up to 800 nm/s, essential for organizing organelles and maintaining cellular architecture.[64] Disruptions in motor protein function impair axonal transport and contribute to neurodegenerative diseases.[65]
