Recent from talks
Contribute something
Nothing was collected or created yet.
Cell (biology)
View on Wikipedia
| Cell | |
|---|---|
 A eukaryotic cell as in animals (left) and a prokaryotic cell as in bacteria (right) | |
| Identifiers | |
| MeSH | D002477 |
| TH | H1.00.01.0.00001 |
| FMA | 686465 |
| Anatomical terminology | |
The cell is the basic structural and functional unit of all forms of life or organisms. The term comes from the Latin word cellula meaning 'small room'. A biological cell basically consists of a semipermeable cell membrane enclosing cytoplasm that contains genetic material. Most cells are only visible under a microscope. Except for highly-differentiated cell types (examples include red blood cells and gametes) most cells are capable of replication, and protein synthesis. Some types of cell are motile. Cells emerged on Earth about four billion years ago.
All organisms are grouped into prokaryotes, and eukaryotes. Prokaryotes are single-celled, and include archaea, and bacteria. Eukaryotes can be single-celled or multicellular, and include protists, plants, animals, most types of fungi, and some species of algae. All multicellular organisms are made up of many different types of cell. The diploid cells that make up the body of a plant or animal are known as somatic cells, and in animals excludes the haploid gametes.
Prokaryotic cells lack the membrane-bound nucleus present in eukaryotic cells, and instead have a nucleoid region. In eukaryotic cells the nucleus is enclosed in the nuclear membrane. Eukaryotic cells contain other membrane-bound organelles such as mitochondria, which provide energy for cell functions, and chloroplasts, in plants that create sugars by photosynthesis. Other non-membrane-bound organelles may be proteinaceous such as the ribosomes present (though different) in both groups. A unique membrane-bound prokaryotic organelle the magnetosome has been discovered in magnetotactic bacteria.
Cells were discovered by Robert Hooke in 1665, who named them after their resemblance to cells in a monastery. Cell theory, developed in 1839 by Matthias Jakob Schleiden and Theodor Schwann, states that all organisms are composed of one or more cells, that cells are the fundamental unit of structure and function in all organisms, and that all cells come from pre-existing cells.
Types
[edit]
Organisms are broadly grouped into eukaryotes, and prokaryotes. Eukaryotic cells possess a membrane-bound nucleus, and prokaryotic cells lack a nucleus but have a nucleoid region.[1] Prokaryotes are single-celled organisms, whereas eukaryotes can be either single-celled or multicellular. Single-celled eukaryotes include microalgae such as diatoms. Multicellular eukaryotes include all animals, and plants, most fungi, and some species of algae.[2][3][4]
Prokaryotes
[edit]
All prokaryotes are single-celled and include bacteria and archaea, two of the three domains of life.[5] Prokaryotic cells were likely the first form of life on Earth,[6][7] characterized by having vital biological processes including cell signaling. They are simpler and smaller than eukaryotic cells, lack a nucleus, and the other usually present membrane-bound organelles.[8] Prokaryotic organelles are simple structures typically non-membrane-bound.[9]

Bacteria
[edit]Bacteria are enclosed in a cell envelope, that protects the interior from the exterior.[10] It generally consists of a plasma membrane covered by a cell wall which, for some bacteria, is covered by a third gelatinous layer called a bacterial capsule. The capsule may be polysaccharide as in pneumococci, meningococci or polypeptide as Bacillus anthracis or hyaluronic acid as in streptococci. Mycoplasma only possess the cell membrane.[11] The cell envelope gives rigidity to the cell and separates the interior of the cell from its environment, serving as a protective mechanical and chemical filter.[12] The cell wall consists of peptidoglycan and acts as an additional barrier against exterior forces.[13][12] The cell wall acts to protect the cell mechanically and chemically from its environment, and is an additional layer of protection to the cell membrane. It also prevents the cell from expanding and bursting (cytolysis) from osmotic pressure due to a hypotonic environment.[14]
The DNA of a bacterium typically consists of a single circular chromosome that is in direct contact with the cytoplasm in a region called the nucleoid. Some bacteria contain multiple circular or even linear chromosomes.[15][16][17] The cytoplasm also contains ribosomes and various inclusions where transcription takes place alongside translation.[18][19] Extrachromosomal DNA as plasmids, are usually circular and encode additional genes, such as those of antibiotic resistance.[20] Linear bacterial plasmids have been identified in several species of spirochete bacteria, including species of Borrelia which causes Lyme disease.[21] The prokaryotic cytoskeleton in bacteria is involved in the maintenance of cell shape, polarity and cytokinesis.[22]
Compartmentalization is a feature of eukaryotic cells but some species of bacteria, have protein-based organelle-like microcompartments such as gas vesicles, and carboxysomes, and encapsulin nanocompartments.[23][24][25][26] Certain membrane-bound prokaryotic organelles have also been discovered. They include the magnetosome of magnetotactic bacteria,[24] and the anammoxosome of anammox bacteria.[27][28]
Cell-surface appendages can include flagella, and pili, protein structures that facilitate movement and communication between cells.[29] The flagellum stretches from the cytoplasm through the cell membrane and extrudes through the cell wall.[30] Fimbriae are short attachment pili, the other type of pilus is the longer conjugative type.[31] Fimbriae are formed of an antigenic protein called pilin, and are responsible for the attachment of bacteria to specific receptors on host cells. [32]
Most prokaryotes are the smallest of all organisms, ranging from 0.5 to 2.0 μm in diameter.[33] The largest bacterium known, Thiomargarita magnifica, is visible to the naked eye with an average length of 1 cm, but can be as much as 2 cm[34] [35]
Archaea
[edit]Archaea are enclosed in a cell envelope consisting of a plasma membrane and a cell wall. An exception to this is the Thermoplasma that only has the cell membrane.[11] The cell membranes of archaea are unique, consisting of ether-linked lipids. The prokaryotic cytoskeleton has homologues of eukaryotic actin and tubulin.[22] A unique form of metabolism in the archaean is methanogenesis. Their cell-surface appendage equivalent of the flagella is the differently structured and unique archaellum.[36][31] The DNA is contained in a circular chromosome in direct contact with the cytoplasm, in a region known as the nucleoid. Ribosomes are also found freely in the cytoplasm, or attached to the cell membrane where DNA processing takes place.[18][37]
The archaea are noted for their extremophile species, and many are selectively evolved to thrive in extreme heat, cold, acidic, alkaline, or high salt conditions.[38] There are no known archaean pathogens.[39]
Eukaryotes
[edit]Eukaryotes can be single-celled, as in diatoms (microscopic algae), or multicellular. Animals, plants, most fungi, and some algae are multicellular.[40] Eukaryotes are distinguished by the presence of a membrane-bound nucleus.[41] The nucleus gives the eukaryote its name, which means "true nut" or "true kernel", where "nut" means the nucleus.[42] Eukaryotic cells can be 2 to 1000 times larger in diameter than a typical prokaryote.[43]
| Property | Archaea | Bacteria | Eukaryota |
|---|---|---|---|
| Cell membrane | Ether-linked lipids | Ester-linked lipids | Ester-linked lipids |
| Cell wall | Glycoprotein, or S-layer; rarely pseudopeptidoglycan | Peptidoglycan, S-layer, or no cell wall | Various structures |
| Gene structure | Circular chromosomes, similar translation and transcription to Eukaryota | Circular chromosomes, unique translation and transcription | Multiple, linear chromosomes, but translation and transcription similar to Archaea |
| Internal cell structure | No membrane-bound organelles (?[44]) or nucleus | No membrane-bound organelles or nucleus | Membrane-bound organelles and nucleus |
| Metabolism[45] | Various, including diazotrophy, with methanogenesis unique to Archaea | Various, including photosynthesis, aerobic and anaerobic respiration, fermentation, diazotrophy, and autotrophy | Photosynthesis, cellular respiration, and fermentation; no diazotrophy |
| Reproduction | Asexual reproduction, horizontal gene transfer | Asexual reproduction, horizontal gene transfer | Sexual and asexual reproduction |
| Protein synthesis initiation | Methionine | Formylmethionine | Methionine |
| RNA polymerase | One | One | Many |
| EF-2/EF-G | Sensitive to diphtheria toxin | Resistant to diphtheria toxin | Sensitive to diphtheria toxin |
Multicellular organisms are made up of many different types of cell known overall as somatic cells.[46] Typical plant cells include parenchyma cells including transfer cells, and collenchyma cells. Animal cells include all those that make up the four main tissue types of epithelium – a number of different epithelial cells; connective tissue such as osteoblasts in bone, and chondrocytes in cartilage; nervous tissue including different brain cells and nerves, and muscle tissue having different muscle cells.[1] The number of cells in these tissues vary with species. Studies on the human have estimated a total cell count at around 30 trillion cells (~36 trillion cells in the male, and ~28 trillion in the female).[47][48]
Eukaryotic cells
[edit]The cells of eukaryotes have a cell membrane that surrounds a gel-like cytoplasm; it contains the cytoskeleton, the cell nucleus, the endomembrane system, and organelles including mitochondria, and the Golgi apparatus.
Cell membrane
[edit]The cell membrane, or plasma membrane, is a selectively permeable membrane as an outer boundary of the cell that encloses the cytoplasm.[49] The membrane serves to separate and protect a cell from its surrounding environment and is made mostly from a lipid bilayer of phospholipids, which are amphiphilic (partly hydrophobic and partly hydrophilic), and is sometimes referred to as a fluid mosaic membrane.[50] Embedded within the cell membrane is a macromolecular structure called the porosome the universal secretory portal in cells and a variety of protein molecules that act as channels and pumps that move different molecules into and out of the cell.[18] The membrane is semi-permeable, and selectively permeable, in that it can either let a substance (molecule or ion) pass through freely, to a limited extent or not at all.[51] Cell surface receptors embedded in the membrane allow cells to detect external signaling molecules such as hormones.[52]
Underlying, and attached to the cell membrane is the cell cortex, the outermost part of the actin cytoskeleton.[53] Its thickness varies with cell type and physiology.
Cytoplasm
[edit]The membrane encloses the cytoplasm of the cell. It is made up of two main components, the cytosol, and the protein filaments that make up the cytoskeleton.[54][55] The cytosol is a gel-like substance made up of water, ions, and non-essential biomolecules. The network of filaments and microtubules of the cytoskeleton gives shape and support to the cell, and has a part in organising the cell components. The cytoplasm surrounds all the organelles of the cell.[54][55]
Cytoskeleton
[edit]The cytoskeleton acts to organize and maintain the cell's shape; anchors organelles in place; helps during endocytosis, the uptake of external materials by a cell, and cytokinesis, the separation of daughter cells after cell division; and moves parts of the cell in processes of growth and mobility. The cytoskeleton is composed of microtubules, intermediate filaments and microfilaments. In a neuron the intermediate filaments are known as neurofilaments. There are a great number of proteins associated with them, each controlling a cell's structure by directing, bundling, and aligning filaments. The outermost part of the cytoskeleton is the cell cortex, or actin cortex, a thin layer of cross-linked actomyosins.[53]
The centrosome is the cytoskeleton organizer in the animal cell that produces the microtubules of a cell—a key component of the cytoskeleton.[56] It directs the transport through the ER and the Golgi apparatus.[57] Centrosomes are composed of two centrioles which lie perpendicular to each other in which each has an organization like a cartwheel, which separate during cell division and help in the formation of the mitotic spindle.[56]
Organelles
[edit]Organelles are parts of the cell that are specialized for carrying out one or more vital functions.[18] There are several types of organelles in a cell held in the gelatinous cytosol of the cytoplasm that fills the cell and surrounds the organelles, forming 30%–50% of a cell volume.[58] Most organelles vary in size and/or number based on the growth of the host cell.[59]
Nucleus
[edit]
The cell nucleus is the largest organelle in the animal cell.[60] It houses the cell's chromosomes, and is the place where almost all DNA replication and RNA synthesis (transcription) occur. The nucleus is spherical and separated from the cytoplasm by a double membrane called the nuclear envelope, space between these two membrane is called perinuclear space. The nuclear envelope isolates and protects a cell's DNA from various molecules that could accidentally damage its structure or interfere with its processing. During processing, DNA is transcribed, or copied into a special RNA, called messenger RNA (mRNA). This mRNA is then transported out of the nucleus, where it is translated into a specific protein molecule. The nucleolus is a specialized region within the nucleus where ribosome subunits are assembled.[18] Cells use DNA for their long-term information storage that is encoded in its DNA sequence.[18] RNA is used for information transport (e.g., mRNA) and enzymatic functions (e.g., ribosomal RNA). Transfer RNA (tRNA) molecules are used to add amino acids during protein translation.[61] Mitochondria have their own DNA (mitochondrial DNA).[62] The mitochondrial genome is a circular DNA molecule distinct from nuclear DNA. Although the mitochondrial DNA is very small compared to nuclear chromosomes,[18] it codes for 13 proteins involved in mitochondrial energy production and specific tRNAs.[63]
The DNA of each cell is its genetic material, and is organized in multiple linear molecules, called chromosomes, that are coiled around histone proteins and housed in the cell nucleus.[41][64] In humans, the nuclear genome is divided into 46 linear chromosomes, including 22 homologous chromosome pairs and a pair of sex chromosomes. The nucleus is a membrane-bound organelle. Other organelles in the cell have specific functions such as mitochondria which provide the cell's energy.[65]
Golgi apparatus
[edit]The Golgi apparatus processes and packages the macromolecules, such as proteins and lipids, that are synthesized by the cell. It is organized as a stack of plate-like structures known as cisternae.[66]
Mitochondria
[edit]Mitochondria generate energy for the cell. Mitochondria are self-replicating double membrane-bound organelles that occur in various numbers, shapes, and sizes in the cytoplasm of the cell.[18] Respiration occurs in the cell mitochondria, which generate the cell's energy by oxidative phosphorylation, using oxygen to release energy stored in cellular nutrients (typically pertaining to glucose) to generate ATP (aerobic respiration).[67] Mitochondria multiply by binary fission.[68]
Lysosomes
[edit]Lysosomes contain enzymes (acid hydrolases). They digest excess or worn-out organelles, food particles, and engulfed viruses or bacteria. Lysosomes are optimally active in an acidic environment. The cell could not house these destructive enzymes if they were not contained in a membrane-bound system.[18][69]
Peroxisomes
[edit]Peroxisomes have enzymes that rid the cell of toxic peroxides,
Vacuoles
[edit]Vacuoles sequester waste products. Some cells, most notably Amoeba, have contractile vacuoles, which can pump water out of the cell if there is too much water.[70]
Endomembrane system
[edit]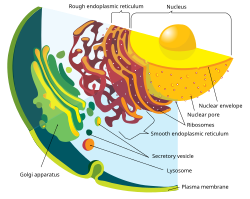
The endomembrane system consists of all the different internal membranes of the cell. These membranes are held in the cell's cytoplasm and divide the various organelles.
Endoplasmic reticulum
[edit]The endoplasmic reticulum (ER) is a transport network for molecules targeted for certain modifications and specific destinations, as compared to molecules that float freely in the cytoplasm. The ER has two forms: the rough endoplasmic reticulum (RER), which has ribosomes on its surface that secrete proteins into the ER, and the smooth endoplasmic reticulum (SER), which lacks ribosomes.[18]
A ribosome is a large complex of RNA and protein molecules.[18] They each consist of two subunits, one larger than the other, and act as an assembly line where RNA from the nucleus is used to synthesise proteins from amino acids. Ribosomes can be found either floating freely or bound to a membrane of the rough endoplasmatic reticulum.[71]
The smooth ER plays a role in calcium sequestration and release, and helps in synthesis of lipid.[72]
Cells of major eukaryote groups
[edit]Animal cells
[edit]
All the cells in an animal body develop from one totipotent diploid cell called a zygote. During the development of an animal, the cells differentiate into the specialised tissues and organs of the organism. (An exception is the simple sponge). The estimated cell count in a typical adult human body is around 30 trillion cells. Different groups of cells differentiate from the three germ layers. Differentiation results in structural or functional changes to the typical eukaryotic cell.
Some types of specialised cell are localised to a particular animal group. Vertebrates for example have specialised, structurally changed cells including muscle cells. The cell membrane of a skeletal muscle cell or of a cardiac muscle cell is termed the sarcolemma.[73] And the cytoplasm is termed the sarcoplasm. Skeletal muscle cells also become multinucleated. Populations of animal groups evolve to become distinct species, where sexual reproduction is isolated. The many species of vertebrates for example have other unique characteristics by way of additional specialised cells. In some species of electric fish for example modified muscle cells or nerve cells have specialised to become electerocytes capable of creating and storing electrical energy for future release, as in stunning prey, or use in electrolocation.[74] These are large flat cells in the electric eel, and electric ray in which thousands are stacked into an electric organ comparable to a voltaic pile.[75]
Organelles are parts of the cell that are specialized for carrying out one or more vital functions, analogous to the organs of the human body (such as the heart, lung, and kidney, with each organ performing a different function).[18] In addition to the organelles shared by all eukaryotes, animal cells often have cilia or flagella.[76][77] Many animal cells are ciliated and most cells except red blood cells have primary cilia. Primary cilia play important roles in chemosensation and mechanosensation.[78] Each cilium may be "viewed as a sensory cellular antennae that coordinates a large number of cellular signaling pathways, sometimes coupling the signaling to ciliary motility or alternatively to cell division and differentiation."[79] Ciliated cells in the respiratory epithelium play an important role in the movement of mucus. Some animal cells have flagella such as flagellated protists and sperm cells, that enable movement.[76] Invertebrate planarians have ciliated excretory flame cells.[80] Other excretory cells also found in planarians are solenocytes that are long and flagellated.
Plant cells
[edit]

Other types of organelle specific to plant cells, are pigment-containing plastids, especially chloroplasts that contain chlorophyll, and large water-storing vacuoles.
Chloroplasts capture the sun's energy to make carbohydrates through photosynthesis.[81]Chromoplasts contain fat-soluble carotenoid pigments such as orange carotene and yellow xanthophylls which helps in synthesis and storage. Leucoplasts are non-pigmented plastids and helps in storage of nutrients.[82] Plastids divide by binary fission. Vacuoles in plant cells store water. They are liquid filled spaces surrounded by a membrane.[83]The vacuoles of plant cells are usually larger than those of animal cells.They are described as liquid filled spaces and are surrounded by a membrane.[83] Vacuoles of plant cells are surrounded by a membrane which transports ions against concentration gradients.[84]
Algal cells
[edit]Algae members are photoautotrophs able to use photosynthesis to produce energy. Photosynthesis is made possible by the use of plastids, organelles in the cytoplasm known as chloroplasts. Algal photoautotrophs include red algae.[85]
Alginate is a polysaccharide found in the matrix of the cell walls of brown algae, and have many important uses in the food industry, and in pharmacology.[86]
Fungal cells
[edit]The cells of fungi have in addition to the shared eukaryotic organelles a spitzenkörper in their endomembrane system, associated with hyphal tip growth. It is a phase-dark body that is composed of an aggregation of membrane-bound vesicles containing cell wall components, serving as a point of assemblage and release of such components intermediate between the Golgi and the cell membrane. The spitzenkörper is motile and generates new hyphal tip growth as it moves forward.[87]
Protist cells
[edit]The cells of protists may be bounded only by a cell membrane, or may in addition have a cell wall, or may be covered by a pellicle (in ciliates), a test (in testate amoebae), or a frustule (in diatoms).
Some protists such as amoebae may feed on other organisms and ingest food by phagocytosis. Vacuoles known as phagosomes in the cytoplasm may be used to draw in and incorporate the captured particles. Other types of protists are photoautotrophs, providing themselves with energy by photosynthesis.[88] Most protists are motile and generate movement with cilia, flagella, or pseudopodia.[88]
Physiology
[edit]
Replication
[edit]
During cell division, part of the cell cycle, a single cell, the mother cell divides into two daughter cells. This leads to growth in multicellular organisms (the growth of tissue) and to procreation (vegetative reproduction) in unicellular organisms. Prokaryotic cells divide by binary fission, while eukaryotic cells usually undergo a process of nuclear division, called mitosis, followed by division of the cell, called cytokinesis. A diploid cell may undergo meiosis to produce haploid cells, usually four. Haploid cells serve as gametes in multicellular organisms, fusing to form new diploid cells.[89]
DNA replication, or the process of duplicating a cell's genome,[18] always happens when a cell divides through mitosis or binary fission.[89] This occurs during the S (synthesis) phase of the cell cycle.
In meiosis, the DNA is replicated only once, while the cell divides twice. DNA replication only occurs before meiosis I. DNA replication does not occur when the cells divide the second time, in meiosis II.[90] Replication, like all cellular activities, requires specialized proteins.[18]
DNA repair
[edit]All cells contain enzyme systems that scan for DNA damage and carry out repair. Diverse repair processes have evolved in all organisms. Repair is vital to maintain DNA integrity, avoid cell death and errors of replication that could lead to mutation. Repair processes include nucleotide excision repair, DNA mismatch repair, non-homologous end joining of double-strand breaks, recombinational repair and light-dependent repair (photoreactivation).[91]
Growth and metabolism
[edit]Between successive cell divisions, cells grow through the functioning of cellular metabolism. Cell metabolism is the process by which individual cells process nutrient molecules. Metabolism has two distinct divisions: catabolism, in which the cell breaks down complex molecules to produce energy and reducing power, and anabolism, in which the cell uses energy and reducing power to construct complex molecules and perform other biological functions.[92]
Complex sugars can be broken down into simpler sugar molecules called monosaccharides such as glucose. Once inside the cell, glucose is broken down to make adenosine triphosphate (ATP),[18] a molecule that possesses readily available energy, through two different pathways. In plant cells, chloroplasts create sugars by photosynthesis, using the energy of light to join molecules of water and carbon dioxide.[93]
Protein synthesis
[edit]Cells are capable of synthesizing new proteins, which are essential for the modulation and maintenance of cellular activities. This process involves the formation of new protein molecules from amino acid building blocks based on information encoded in DNA/RNA. Protein synthesis generally consists of two major steps: transcription and translation.[61]
Transcription is the process where genetic information in DNA is used to produce a complementary RNA strand. This RNA strand is then processed to give messenger RNA (mRNA), which is free to migrate into the cytoplasm. mRNA molecules bind to protein-RNA complexes called ribosomes located in the cytosol, where they are translated into polypeptide sequences. The ribosome mediates the formation of a polypeptide sequence based on the mRNA sequence. The mRNA sequence directly relates to the polypeptide sequence by binding to transfer RNA (tRNA) adapter molecules in binding pockets within the ribosome.[61] The new polypeptide then folds into a functional three-dimensional protein molecule.
Motility
[edit]Unicellular organisms can move in order to find food or escape predators. Common mechanisms of motion include flagella and cilia.[31]
In multicellular organisms, cells can move during processes such as wound healing, the immune response and cancer metastasis. For example, in wound healing in animals, white blood cells move to the wound site to kill the microorganisms that cause infection. Cell motility involves many receptors, crosslinking, bundling, binding, adhesion, motor and other proteins.[94] The process is divided into three steps: protrusion of the leading edge of the cell, adhesion of the leading edge and de-adhesion at the cell body and rear, and cytoskeletal contraction to pull the cell forward. Each step is driven by physical forces generated by unique segments of the cytoskeleton.[95][94]
Navigation, control and communication
[edit]In August 2020, scientists described one way cells—in particular cells of a slime mold and mouse pancreatic cancer-derived cells—are able to navigate efficiently through a body and identify the best routes through complex mazes: generating gradients after breaking down diffused chemoattractants which enable them to sense upcoming maze junctions before reaching them, including around corners.[96][97][98]
Death
[edit]Cell death is the event of a biological cell ceasing to carry out its functions. This may be the result of the natural process of old cells dying and being replaced by new ones, as in programmed cell death, or may result from factors such as diseases, localized injury, exposure to a toxic substance, or the death of the host organism. Apoptosis or Type I cell-death, and autophagy or Type II cell-death are both forms of programmed cell death, while necrosis is a non-physiological process that occurs as a result of infection or injury.[99][100]
Cell ancestry traces back in an unbroken lineage for over 3 billion years. The immortality of a cell lineage depends on the maintenance of cell division potential,[101] which may be lost because of cell damage, terminal differentiation as occurs in nerve cells, or programmed cell death during development. Maintenance of division potential over successive generations depends on the avoidance and the accurate repair of cellular damage, particularly DNA damage. Sexual processes provide an opportunity for effective repair of DNA damage in the germ line by homologous recombination.[101][102]
Multicellularity
[edit]Cell differentiation
[edit]
Multicellular organisms are organisms that consist of more than one cell, in contrast to single-celled organisms.[103] Microorganisms cloned from a single cell can form visible microbial colonies. A microbial consortium of two or more species of microorganisms can form a biofilm community,[104] such as dental plaque. The cell-to-cell adhesion found in microbial colonies may have been the first evolutionary step toward more complex multicellular organisms.[105]
In complex multicellular organisms, cells specialize into different cell types that are adapted to particular functions.[106] In animals, major cell types include skin cells, muscle cells, neurons, blood cells, fibroblasts, stem cells, and others. Cell types differ both in appearance and function, yet are genetically identical. Cells are able to be of the same genotype but of different cell type due to the differential expression of the genes they contain.[107]
Most distinct cell types arise from a single totipotent cell, called a zygote, that differentiates into hundreds of different cell types during the course of development. Differentiation of cells is driven by different environmental cues (such as cell–cell interaction) and intrinsic differences (such as those caused by the uneven distribution of molecules during division).[108]
Signaling
[edit]Cell signaling is the process by which a cell interacts with itself, other cells, and the environment. Typically, the signaling process involves three components: the first messenger (the ligand), the receptor, and the signal itself.[109] Most cell signaling is chemical in nature, and can occur with neighboring cells or more distant targets. Signal receptors are complex proteins or tightly bound multimer of proteins, located in the plasma membrane or within the interior.[110]
Each cell is programmed to respond to specific extracellular signal molecules, and this process is the basis of development, tissue repair, immunity, and homeostasis. Individual cells are able to manage receptor sensitivity including turning them off, and receptors can become less sensitive when they are occupied for long durations.[110] Errors in signaling interactions may cause diseases such as cancer, autoimmunity, and diabetes.[111]
Origin of multicellularity
[edit]Multicellularity has evolved independently at least 25 times,[112] including in some prokaryotes, like cyanobacteria, myxobacteria, actinomycetes, or Methanosarcina. However, complex multicellular organisms evolved only in six eukaryotic groups: animals, fungi, brown algae, red algae, green algae, and plants.[113] It evolved repeatedly for plants (Chloroplastida), once or twice for animals, once for brown algae, and perhaps several times for fungi, slime molds, and red algae.[114] Multicellularity may have evolved from colonies of interdependent organisms, from cellularization, or from organisms in symbiotic relationships.[115]
The first evidence of multicellularity is from cyanobacteria-like organisms that lived between 3 and 3.5 billion years ago.[112] Other early fossils of multicellular organisms include the contested Grypania spiralis and the fossils of the black shales of the Palaeoproterozoic Francevillian Group Fossil B Formation in Gabon.[116]
The evolution of multicellularity from unicellular ancestors has been replicated in the laboratory, in evolution experiments using predation as the selective pressure.[112]
Origins
[edit]The origin of cells has to do with the origin of life, which began the history of life on Earth.
Origin of life
[edit]
Small molecules needed for life may have been carried to Earth on meteorites, created at deep-sea vents, or synthesized by lightning in a reducing atmosphere. There is little experimental data defining what the first self-replicating forms were. RNA may have been the earliest self-replicating molecule, as it can both store genetic information and catalyze chemical reactions.[117] This process required an enzyme to catalyze the RNA reactions, which may have been the early peptides that formed in hydrothermal vents.[118]
Cells emerged around 4 billion years ago.[119][120] The first cells were most likely heterotrophs. The early cell membranes were probably simpler and more permeable than modern ones, with only a single fatty acid chain per lipid. Lipids spontaneously form bilayered vesicles in water, and could have preceded RNA.[121][122]
First eukaryotes
[edit]
Eukaryotic cells were created some 2.2 billion years ago in a process called eukaryogenesis. This is widely agreed to have involved symbiogenesis, in which archaea and bacteria came together to create the first eukaryotic common ancestor.[123] However, the sequence of the steps involved has been disputed.[citation needed] It evolved into a population of single-celled organisms that included the last eukaryotic common ancestor, gaining capabilities along the way.[124][125]
This cell had a new level of complexity and capability, with a nucleus[126][124] and facultatively aerobic mitochondria.[123] It featured at least one centriole and cilium, sex (meiosis and syngamy), peroxisomes, and a dormant cyst with a cell wall of chitin and/or cellulose.[127][125] The last eukaryotic common ancestor gave rise to the eukaryotes' crown group, containing the ancestors of animals, fungi, plants, and a diverse range of single-celled organisms.[128][129] The plants were created around 1.6 billion years ago with a second episode of symbiogenesis that added chloroplasts, derived from cyanobacteria.[123]
History of research
[edit]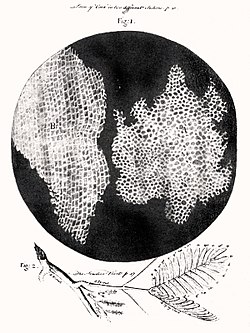
In 1665, Robert Hooke examined a thin slice of cork under his microscope, and saw a structure of small enclosures. He wrote "I could exceeding plainly perceive it to be all perforated and porous, much like a honeycomb, but that the pores of it were not regular".[130] To further support his theory, Matthias Schleiden and Theodor Schwann studied cells of both animal and plants. What they discovered were significant differences between the two types of cells. This put forth the idea that cells were fundamental to both plants and animals.[131]
- 1632–1723: Antonie van Leeuwenhoek taught himself to make lenses, constructed basic optical microscopes and drew protozoa, such as Vorticella from rain water, and bacteria from his own mouth.[132]
- 1665: Robert Hooke discovered cells in cork, then in living plant tissue using an early microscope. In his book Micrographia he coined the term cell (from Latin cellula, meaning "small room") since they resembled the cells of a monastery[133][134][135][136][132]
- 1839: Theodor Schwann[137] and Matthias Jakob Schleiden elucidated the principle that plants and animals are made of cells, concluding that cells are a common unit of structure and development, founding the cell theory.[138][139]
- 1855: Rudolf Virchow stated that new cells come from pre-existing cells by cell division (omnis cellula ex cellula).
- 1931: Ernst Ruska built the first transmission electron microscope at the University of Berlin.[140] By 1935, he had built an electron microscope with twice the resolution of a light microscope, revealing previously unresolvable organelles.
- 1981: Lynn Margulis published Symbiosis in Cell Evolution detailing how eukaryotic cells were created by symbiogenesis.[141]
See also
[edit]References
[edit]- ^ a b Cooper, Geoffrey M. (2000). "The Origin and Evolution of Cells". The Cell: A Molecular Approach. 2nd edition. Sinauer Associates. Retrieved 17 September 2025.
- ^ "Prokaryote structure". khanacademy. Retrieved 2025-08-16.
- ^ Knoll, Andrew H. (2011). "The Multiple Origins of Complex Multicellularity". Annual Review of Earth and Planetary Sciences. 39: 217–239. Bibcode:2011AREPS..39..217K. doi:10.1146/annurev.earth.031208.100209.
- ^ "24.1B: Fungi Cell Structure and Function". Biology LibreTexts. 15 July 2018. Retrieved 3 October 2025.
- ^ Cole, Laurence A. (2016-01-01), Cole, Laurence A. (ed.), "Chapter 13 - Evolution of Chemical, Prokaryotic, and Eukaryotic Life", Biology of Life, Academic Press, pp. 93–99, doi:10.1016/b978-0-12-809685-7.00013-7, ISBN 978-0-12-809685-7, retrieved 2025-08-16
- ^ "Evolutionary History of Prokaryotes". courses.lumenlearning.com. Biology for Majors II. Retrieved 2025-08-16.
- ^ Poole, Anthony; Jeffares, Daniel; Penny, David (1999). "Early evolution: prokaryotes, the new kids on the block". BioEssays. 21 (10): 880–889. doi:10.1002/(SICI)1521-1878(199910)21:10<880::AID-BIES11>3.0.CO;2-P. ISSN 1521-1878. PMID 10497339.
- ^ Fowler, Samantha; Roush, Rebecca; Wise, James (2013-04-25). "3.2 Comparing Prokaryotic and Eukaryotic Cells - Concepts of Biology | OpenStax". openstax.org. Retrieved 2025-08-22.
- ^ Grant, Carly R.; et al. (October 2018). "Organelle Formation in Bacteria and Archaea". Annual Review of Cell and Developmental Biology. 34: 217–238. doi:10.1146/annurev-cellbio-100616-060908. PMID 30113887.
- ^ Silhavy, Thomas J.; Kahne, Daniel; Walker, Suzanne (2010-05-01). "The Bacterial Cell Envelope". Cold Spring Harbor Perspectives in Biology. 2 (5) a000414. doi:10.1101/cshperspect.a000414. ISSN 1943-0264. PMC 2857177. PMID 20452953.
- ^ a b Barton, Larry L. (2005). Structural and Functional Relationships in Prokaryotes. SpringerLink: Springer e-Books. Springer Science & Business Media. pp. 69–71. ISBN 978-0-387-27125-5.
- ^ a b Fuertes-Rabanal, María; et al. (July 2, 2025). "Cell walls: a comparative view of the composition of cell surfaces of plants, algae, and microorganisms". Journal of Experimental Botany. 76 (10): 2614–2645. doi:10.1093/jxb/erae512. PMC 12223506. PMID 39705009.
- ^ Seltmann, Guntram; Holst, Otto (2013). The Bacterial Cell Wall. Springer Science & Business Media. p. 3. ISBN 978-3-662-04878-8.
- ^ Prasad, Krishna Kant; Prasad, Nooralabettu Krishna (2010). Downstream Process Technology: A New Horizon In Biotechnology. PHI Learning Pvt. Ltd. pp. 116–117. ISBN 978-81-203-4040-4.
- ^ Egan, Elizabeth S.; Fogel, Michael A.; Waldor, Matthew K. (2005). "MicroReview: Divided genomes: negotiating the cell cycle in prokaryotes with multiple chromosomes". Molecular Microbiology. 56 (5): 1129–1138. doi:10.1111/j.1365-2958.2005.04622.x. PMID 15882408.
- ^ "Genome Packaging in Prokaryotes | Learn Science at Scitable". www.nature.com. Retrieved 2025-08-30.
- ^ "The difference between nucleus and nucleoid". BYJUS. Retrieved 2025-08-30.
- ^ a b c d e f g h i j k l m n o
 This article incorporates public domain material from "What Is a Cell?". Science Primer. NCBI. 30 March 2004. Archived from the original on 2009-12-08. Retrieved 3 May 2013.
This article incorporates public domain material from "What Is a Cell?". Science Primer. NCBI. 30 March 2004. Archived from the original on 2009-12-08. Retrieved 3 May 2013.
- ^ "7.6C: Prokaryotic Transcription and Translation Are Coupled". Biology LibreTexts. 17 May 2017. Retrieved 17 October 2025.
- ^ Henkin, Tina M.; Peters, Joseph E. (2020). Snyder and Champness Molecular Genetics of Bacteria. ASM Books (5th ed.). John Wiley & Sons. pp. 181–189. ISBN 978-1-55581-975-0.
- ^ "Karyn's Genomes: Borrelia burgdorferi". 2can on the EBI-EMBL database. European Bioinformatics Institute. Archived from the original on 2013-05-06. Retrieved 2013-05-06.
- ^ a b Erickson HP (February 2017). "The discovery of the prokaryotic cytoskeleton: 25th anniversary". Mol Biol Cell. 28 (3): 357–358. doi:10.1091/mbc.E16-03-0183. PMC 5341718. PMID 28137947.
- ^ McDowell HB, Hoiczyk E (March 2022). "Bacterial Nanocompartments: Structures, Functions, and Applications". J Bacteriol. 204 (3) e00346-21: e0034621. doi:10.1128/JB.00346-21. PMC 8923211. PMID 34606372.
- ^ a b Murat, D; Byrne, M; Komeili, A (October 2010). "Cell biology of prokaryotic organelles". Cold Spring Harbor Perspectives in Biology. 2 (10) a000422. doi:10.1101/cshperspect.a000422. PMC 2944366. PMID 20739411.
- ^ Stewart, Katie L.; Stewart, Andrew M.; Bobik, Thomas A. (2020-10-06). "Prokaryotic Organelles: Bacterial Microcompartments in E. coli and Salmonella". EcoSal Plus. 9 (1) 10.1128/ecosalplus.ESP–0025–2019. doi:10.1128/ecosalplus.esp-0025-2019. PMC 7552817. PMID 33030141.
- ^ Adamiak N, Krawczyk KT, Locht C, Kowalewicz-Kulbat M (2021). "Archaeosomes and Gas Vesicles as Tools for Vaccine Development". Front Immunol. 12 746235. doi:10.3389/fimmu.2021.746235. PMC 8462270. PMID 34567012.
- ^ de Almeida, NM; Neumann, S; Mesman, RJ; Ferousi, C; Keltjens, JT; Jetten, MS; Kartal, B; van Niftrik, L (July 2015). "Immunogold Localization of Key Metabolic Enzymes in the Anammoxosome and on the Tubule-Like Structures of Kuenenia stuttgartiensis". Journal of Bacteriology. 197 (14): 2432–41. doi:10.1128/JB.00186-15. PMC 4524196. PMID 25962914.
- ^ Saier Jr., Milton H.; Bogdanov, Mikhail V. (2013). "Membranous Organelles in Bacteria". Microbial Physiology. 23 (1–2): 5–12. doi:10.1159/000346496. PMID 23615191.
- ^ Kim, K. W. (2017). "Electron microscopic observations of prokaryotic surface appendages". Journal of Microbiology. 55 (12): 919–926. doi:10.1007/s12275-017-7369-4. PMID 29214488.
- ^ Cohen-Bazire, Germaine; London, Jack (August 1967). "Basal Organelles of Bacterial Flagella". Journal of Bacteriology. 94 (2): 458–465. doi:10.1128/jb.94.2.458-465.1967. PMC 315060. PMID 6039362.
- ^ a b c Beeby, Morgan; et al. (May 2020). "Propulsive nanomachines: the convergent evolution of archaella, flagella and cilia". FEMS Microbiology Reviews. 44 (3): 253–304. doi:10.1093/femsre/fuaa006. PMID 32149348.
- ^ Wilson, Brenda A.; et al. (2020). Bacterial Pathogenesis: A Molecular Approach. ASM Books (4th ed.). John Wiley & Sons. p. 270. ISBN 978-1-55581-941-5.
- ^ Black, Jacquelyn G. (2004). Microbiology. New York Chichester: Wiley. p. 78. ISBN 978-0-471-42084-2.
- ^ Volland, Jean-Marie; et al. (June 24, 2022). "A centimeter-long bacterium with DNA contained in metabolically active, membrane-bound organelles". Science. 376 (6600): 1453–1458. Bibcode:2022Sci...376.1453V. bioRxiv 10.1101/2022.02.16.480423. doi:10.1126/science.abb3634. eISSN 1095-9203. ISSN 0036-8075. PMID 35737788. S2CID 249990020.
- ^ Pennisi, Elizabeth. "Largest bacterium ever discovered has unexpectedly complex cells". Science. science.org. Retrieved 2022-02-24.
- ^ van Wolferen, Marleen; Pulschen, Andre Arashiro; Baum, Buzz; Gribaldo, Simonetta; Albers, Sonja-Verena (November 2022). "The cell biology of archaea". Nature Microbiology. 7 (11): 1744–1755. doi:10.1038/s41564-022-01215-8. ISSN 2058-5276. PMC 7613921. PMID 36253512.
- ^ Ménétret, Jean-François; Schaletzky, Julia; Clemons, William M.; et al. (December 2007). "Ribosome binding of a single copy of the SecY complex: implications for protein translocation" (PDF). Molecular Cell. 28 (6): 1083–1092. doi:10.1016/j.molcel.2007.10.034. PMID 18158904. Archived (PDF) from the original on 2021-01-21. Retrieved 2020-09-01.
- ^ Rampelotto, Pabulo Henrique (2013). "Extremophiles and Extreme Environments". Life. 3 (3): 482–485. Bibcode:2013Life....3..482R. doi:10.3390/life3030482. PMC 4187170. PMID 25369817.
- ^ Duller S, Moissl-Eichinger C (August 2024). "Archaea in the Human Microbiome and Potential Effects on Human Infectious Disease". Emerg Infect Dis. 30 (8): 1505–13. doi:10.3201/eid3008.240181. PMC 11286065. PMID 39043386.
- ^ Lodé, Thierry (2012-10-19). "For Quite a Few Chromosomes More: The Origin of Eukaryotes…". Journal of Molecular Biology. 423 (2): 135–142. doi:10.1016/j.jmb.2012.07.005. ISSN 0022-2836. PMID 22796299.
- ^ a b Visible Body, part of Cengage Learning. "Eukaryotic Chromosomes". www.visiblebody.com. Retrieved 2025-09-11.
- ^ "More on Eukaryote Morphology". ucmp.berkeley.edu. Retrieved 2025-09-03.
- ^ Bartee, Lisa; Shriner, Walter; Creech, Catherine (2017). "Comparing Prokaryotic and Eukaryotic Cells". OpenOregon Educational Resources.
- ^ Heimerl T, Flechsler J, Pickl C, Heinz V, Salecker B, Zweck J, Wanner G, Geimer S, Samson RY, Bell SD, Huber H, Wirth R, Wurch L, Podar M, Rachel R (13 June 2017). "A Complex Endomembrane System in the Archaeon Ignicoccus hospitalis Tapped by Nanoarchaeum equitans". Frontiers in Microbiology. 8 1072. doi:10.3389/fmicb.2017.01072. PMC 5468417. PMID 28659892.
- ^ Jurtshuk P (1996). "Bacterial Metabolism". Medical Microbiology (4th ed.). Galveston (TX): University of Texas Medical Branch at Galveston. ISBN 978-0-9631172-1-2. PMID 21413278.
- ^ "Somatic Cells". www.genome.gov. Retrieved 19 September 2025.
- ^ Hatton, Ian A.; Galbraith, Eric D.; Merleau, Nono S. C.; Miettinen, Teemu P.; Smith, Benjamin McDonald; Shander, Jeffery A. (2023-09-26). "The human cell count and size distribution". Proceedings of the National Academy of Sciences. 120 (39) e2303077120. Bibcode:2023PNAS..12003077H. doi:10.1073/pnas.2303077120. ISSN 0027-8424. PMC 10523466. PMID 37722043.
- ^ "Human Cell data". humancelltreemap.mis.mpg.de. Retrieved 23 September 2025.
- ^ "Inside the Cell" (PDF). publications.nigms.nih.gov. Archived from the original (PDF) on 28 July 2017. Retrieved 22 September 2025.
- ^ Singer, S. J.; Nicolson, Garth L. (February 18, 1972). "The Fluid Mosaic Model of the Structure of Cell Membranes: Cell membranes are viewed as two-dimensional solutions of oriented globular proteins and lipids". Science. 175 (4023): 720–731. doi:10.1126/science.175.4023.720. PMID 4333397.
- ^ Stillwell, William (April 26, 2013). "Membrane Transport". An Introduction to Biological Membranes. pp. 305–337. doi:10.1016/B978-0-444-52153-8.00014-3. ISBN 978-0-444-52153-8. PMC 7182113.
- ^ Guyton, Arthur C.; Hall, John E. (2016). Guyton and Hall Textbook of Medical Physiology. Philadelphia: Elsevier Saunders. pp. 930–937. ISBN 978-1-4557-7005-2. OCLC 1027900365.
- ^ a b Zhu H, Miao R, Wang J, Lin M (March 2024). "Advances in modeling cellular mechanical perceptions and responses via the membrane-cytoskeleton-nucleus machinery". Mechanobiol Med. 2 (1) 100040. doi:10.1016/j.mbm.2024.100040. PMC 12082147. PMID 40395451.
- ^ a b "Cytoplasm, Cytosol and Cytoskeleton". Journal of Clinical Research and Medicine. 5 (5). 28 September 2022. doi:10.31038/JCRM.2022552.
- ^ a b Alberts, Bruce (2015). Molecular biology of the cell (6th ed.). New York: Garland science, Taylor and Francis group. p. 642. ISBN 978081534464-3.
- ^ a b Prigent, Claude; Uzbekov, Rustem (2022). "Duplication and Segregation of Centrosomes during Cell Division". Cells. 11 (15) 2445. doi:10.3390/cells11152445. PMC 9367774. PMID 35954289.
- ^ Gurel, Pinar S.; et al. (July 21, 2014). "Connecting the Cytoskeleton to the Endoplasmic Reticulum and Golgi". Current Biology. 24 (14): R660 – R672. doi:10.1016/j.biochi.2015.03.021. PMC 4678951. PMID 25869000.
- ^ Vekilov, P. G.; et al. (2008). "Metastable mesoscopic phases in concentrated protein solutions". In Franzese, Giancarlo; Rubi, Miguel (eds.). Aspects of Physical Biology: Biological Water, Protein Solutions, Transport and Replication. Lecture Notes in Physics. Vol. 752. Springer Science & Business Media. pp. 65–66. ISBN 978-3-540-78764-8.
- ^ Marshall, Wallace F. (2020). "Scaling of Subcellular Structures". Annual Review of Cell and Developmental Biology. 36: 219–236. doi:10.1146/annurev-cellbio-020520-113246. PMC 8562892. PMID 32603615.
- ^ Khan, Yusuf S.; Farhana, Aisha (2025). "Histology, Cell". StatPearls. StatPearls Publishing. Retrieved 15 October 2025.
- ^ a b c Colville, Thomas P.; Bassert, Joanna M. (2015). Clinical Anatomy and Physiology for Veterinary Technicians (3rd ed.). Elsevier Health Sciences. pp. 93–95. ISBN 978-0-323-22793-3.
- ^ Allen, John F. (February 14, 2003). "Why chloroplasts and mitochondria contain genomes". Comparative and Functional Genomics. 4 (1): 31–36. doi:10.1002/cfg.245. PMC 2447392. PMID 18629105.
- ^ Boore, Jeffrey L. (April 1999). "Animal mitochondrial genomes Open Access". Nucleic Acids Research. 27 (8): 1767–1780. doi:10.1093/nar/27.8.1767. PMC 148383. PMID 10101183.
- ^ Visible Body, part of Cengage Learning. "Eukaryotic vs. Prokaryotic Chromosomes". www.visiblebody.com. Retrieved 2025-09-11.
- ^ Gabaldón, Toni; Pittis, Alexandros A. (2015). "Origin and evolution of metabolic sub-cellular compartmentalization in eukaryotes". Biochimie. 119: 262–268. doi:10.1016/j.biochi.2015.03.021. PMC 4678951. PMID 25869000.
- ^ Short, Ben; Barr, Francis A. (August 14, 2000). "The Golgi apparatus". Current Biology. 10 (16): R583 – R585. Bibcode:2000CBio...10.R583S. doi:10.1016/S0960-9822(00)00644-8. PMID 10985372.
- ^ Beard, Daniel A. (September 9, 2005). "A Biophysical Model of the Mitochondrial Respiratory System and Oxidative Phosphorylation". PLOS Computational Biology. 1 (4): e36. Bibcode:2005PLSCB...1...36B. doi:10.1371/journal.pcbi.0010036. PMC 1201326. PMID 16163394.
- ^ Steven, Alasdair; et al. (2016). Molecular Biology of Assemblies and Machines. Garland Science, Taylor & Francis Group. ISBN 978-1-134-98282-0.
- ^ Soto, Ubaldo; et al. (1993). "Peroxisomes and Lysosomes". In LeBouton, Albert V. (ed.). Molecular & Cell Biology of the Liver. CRC Press. pp. 181–211. ISBN 978-0-8493-8891-0.
- ^ Pappas, George D.; Brandt, Philip W. (1958). "The Fine Structure of the Contractile Vacuole in Ameba". The Journal of Biophysical and Biochemical Cytology. 4 (4): 485–488. doi:10.1083/jcb.4.4.485. ISSN 0095-9901. JSTOR 1603216. PMC 2224495. PMID 13563556.
- ^ Ménétret, Jean-François; Schaletzky, Julia; Clemons, William M.; et al. (December 2007). "Ribosome binding of a single copy of the SecY complex: implications for protein translocation" (PDF). Molecular Cell. 28 (6): 1083–1092. doi:10.1016/j.molcel.2007.10.034. PMID 18158904. Archived (PDF) from the original on 2021-01-21. Retrieved 2020-09-01.
- ^ Pavelka, M.; Roth, J. (2010). "Smooth Endoplasmic Reticulum". Functional Ultrastructure. Vienna: Springer. pp. 42–43. doi:10.1007/978-3-211-99390-3_23. ISBN 978-3-211-99389-7.
- ^ Roberts, MD; Haun, CT; Vann, CG; Osburn, SC; Young, KC (2020). "Sarcoplasmic Hypertrophy in Skeletal Muscle: A Scientific "Unicorn" or Resistance Training Adaptation?". Frontiers in Physiology. 11 816. doi:10.3389/fphys.2020.00816. PMC 7372125. PMID 32760293.
- ^ Markham MR (July 2013). "Electrocyte physiology: 50 years later". J Exp Biol. 216 (Pt 13): 2451–8. Bibcode:2013JExpB.216.2451M. doi:10.1242/jeb.082628. PMID 23761470.
- ^ Mauro A (April 1969). "The role of the Voltaic pile in the Galvani-Volta controversy concerning animal vs. metallic electricity". J Hist Med Allied Sci. 24 (2): 140–50. doi:10.1093/jhmas/xxiv.2.140. PMID 4895861.
- ^ a b Mitchell, D. R. (2007). "The evolution of eukaryotic cilia and flagella as motile and sensory organelles". Eukaryotic Membranes and Cytoskeleton. Adv Exp Med Biology. Vol. 607. pp. 130–40. doi:10.1007/978-0-387-74021-8_11. ISBN 978-0-387-74020-1. PMC 3322410. PMID 17977465.
- ^ Zhang, Qing; Hu, Jinghua; Ling, Kun (2013-09-10). "Molecular views of Arf-like small GTPases in cilia and ciliopathies". Experimental Cell Research. Special Issue: Small GTPases. 319 (15): 2316–2322. doi:10.1016/j.yexcr.2013.03.024. ISSN 0014-4827. PMC 3742637. PMID 23548655.
- ^ Haycraft, Courtney J.; Serra, Rosa (2008-01-01), "Chapter 11 Cilia Involvement in Patterning and Maintenance of the Skeleton", Current Topics in Developmental Biology, Ciliary Function in Mammalian Development, 85, Academic Press: 303–332, doi:10.1016/s0070-2153(08)00811-9, ISBN 978-0-12-374453-1, PMC 3107512, PMID 19147010
- ^ Satir, P.; Christensen, Søren T. (June 2008). "Structure and function of mammalian cilia". Histochemistry and Cell Biology. 129 (6): 687–693. doi:10.1007/s00418-008-0416-9. PMC 2386530. PMID 18365235. 1432-119X.
- ^ "41.8: Excretion Systems - Flame Cells of Planaria and Nephridia of Worms". Biology LibreTexts. 17 July 2018. Retrieved 18 October 2025.
- ^ Björn, Lars Olof; Govindjee (2009). "The evolution of photosynthesis and chloroplasts". Current Science. 96 (11 June 10, 2009): 1466–1474. JSTOR 24104775.
- ^ Sato, N. (2006). "Origin and Evolution of Plastids: Genomic View on the Unification and Diversity of Plastids". In Wise, R. R.; Hoober, J. K. (eds.). The Structure and Function of Plastids. Advances in Photosynthesis and Respiration. Vol. 23. Springer. pp. 75–102. doi:10.1007/978-1-4020-4061-0_4. ISBN 978-1-4020-4060-3.
- ^ a b Lew, Kristi; Fitzpatrick, Brad (2021). Plant Cells (Third ed.). Infobase Holdings, Inc. pp. 14–16. ISBN 978-1-64693-728-8.
- ^ Etxeberria, Ed; et al. (July 2012). "In and out of the plant storage vacuole". Plant Science. 190: 52–61. Bibcode:2012PlnSc.190...52E. doi:10.1016/j.plantsci.2012.03.010. PMID 22608519.
- ^ Yoon, Hwan Su; Hackett, Jeremiah D.; Ciniglia, Claudia; Pinto, Gabriele; Bhattacharya, Debashish (May 2004). "A Molecular Timeline for the Origin of Photosynthetic Eukaryotes". Molecular Biology and Evolution. 21 (5): 809–818. doi:10.1093/molbev/msh075. ISSN 1537-1719. PMID 14963099.
- ^ Abka-Khajouei R, Tounsi L, Shahabi N, Patel AK, Abdelkafi S, Michaud P (May 2022). "Structures, Properties and Applications of Alginates". Mar Drugs. 20 (6): 364. doi:10.3390/md20060364. PMC 9225620. PMID 35736167.
- ^ Steinberg G (March 2007). "Hyphal growth: a tale of motors, lipids, and the Spitzenkörper". Eukaryotic Cell. 6 (3): 351–60. doi:10.1128/EC.00381-06. PMC 1828937. PMID 17259546.
- ^ a b "8.16E: Cell Structure, Metabolism, and Motility". Biology LibreTexts. 23 June 2017. Retrieved 14 October 2025.
- ^ a b Vega, Leslie; White, Bret (2019). Fundamentals of Genetics. Ed-tech Press. pp. 147–148. ISBN 978-1-83947-450-7.
- ^ Campbell Biology – Concepts and Connections. Pearson Education. 2009. p. 138.
- ^ Snustad, D. Peter; Simmons, Michael J. (2015). "DNA repair mechanisms". Principles of Genetics (7th ed.). John Wiley & Sons. pp. 333–336. ISBN 978-1-119-14228-7.
- ^ Smolin, Lori A.; Grosvenor, Mary B. (2019). Nutrition: Science and Applications (4 ed.). John Wiley & Sons. pp. 99–100. ISBN 978-1-119-49527-7.
- ^ Alberts, B.; et al. (2002). "Chloroplasts and Photosynthesis". Molecular Biology of the Cell (4th ed.). New York: Garland Science. Retrieved 2025-10-04.
- ^ a b Ananthakrishnan, R.; Ehrlicher, A. (June 2007). "The forces behind cell movement". International Journal of Biological Sciences. 3 (5). Biolsci.org: 303–317. doi:10.7150/ijbs.3.303. PMC 1893118. PMID 17589565.
- ^ Alberts, Bruce (2002). Molecular biology of the cell (4th ed.). Garland Science. pp. 973–975. ISBN 0-8153-4072-9.
- ^ Willingham, Emily. "Cells Solve an English Hedge Maze with the Same Skills They Use to Traverse the Body". Scientific American. Archived from the original on 4 September 2020. Retrieved 7 September 2020.
- ^ "How cells can find their way through the human body". phys.org. Archived from the original on 3 September 2020. Retrieved 7 September 2020.
- ^ Tweedy, Luke; Thomason, Peter A.; Paschke, Peggy I.; Martin, Kirsty; Machesky, Laura M.; Zagnoni, Michele; Insall, Robert H. (August 2020). "Seeing around corners: Cells solve mazes and respond at a distance using attractant breakdown". Science. 369 (6507) eaay9792. doi:10.1126/science.aay9792. PMID 32855311. S2CID 221342551. Archived from the original on 2020-09-12. Retrieved 2020-09-13.
- ^ D'Arcy, Mark S. (June 2019). "Cell death: a review of the major forms of apoptosis, necrosis and autophagy". Cell Biology International. 43 (6): 582–592. doi:10.1002/cbin.11137. PMID 30958602.
- ^ Yuan, J.; Ofengeim, D. (2024). "A guide to cell death pathways". Nature Reviews Molecular Cell Biology. 25 (5): 379–395. doi:10.1038/s41580-023-00689-6. PMID 38110635.
- ^ a b Bernstein, C.; et al. (2000). "Cell Immortality: Maintenance of Cell Division Potential". Cell Immortalization. Progress in Molecular and Subcellular Biology. Vol. 24. pp. 23–50. doi:10.1007/978-3-662-06227-2_2. ISBN 978-3-642-08491-1. PMID 10547857.
- ^ Avise, J. C. (October 1993). "Perspective: The evolutionary biology of aging, sexual reproduction, and DNA repair". Evolution. 47 (5): 1293–1301. Bibcode:1993Evolu..47.1293A. doi:10.1111/j.1558-5646.1993.tb02155.x. PMID 28564887.
- ^ Becker, Wayne M.; et al. (2009). The world of the cell. Pearson Benjamin Cummings. p. 480. ISBN 978-0-321-55418-5.
- ^ Pátková, Irena; et al. (2012). "Developmental plasticity of bacterial colonies and consortia in germ-free and gnotobiotic settings". BMC Microbiology. 12 178. doi:10.1186/1471-2180-12-178. PMC 3583141. PMID 22894147.
- ^ Niklas, Karl J.; Newman, Stuart A. (January 2013). "The origins of multicellular organisms". Evolution & Development. 15 (1): 41–52. doi:10.1111/ede.12013. PMID 23331916.
- ^ Zeng, Hongkui (2022). "What is a cell type and how to define it?". Cell. 185 (15): 2739–2755. doi:10.1016/j.cell.2022.06.031. ISSN 0092-8674. PMC 9342916. PMID 35868277.
- ^ Aruni, A. Wilson; Ramadass, P. (2019). Animal Tissue Culture. MJP Publisher. pp. 167–169. ISBN 978-81-8094-056-9.
- ^ Deasy, Bridget M. (2009). "Asymmetric Behavior in Stem Cells". In Rajasekhar, V. K.; Vemuri, M. C. (eds.). Regulatory Networks in Stem Cells. Stem Cell Biology and Regenerative Medicine. Humana Press. pp. 13–22. doi:10.1007/978-1-60327-227-8_2. ISBN 978-1-60327-227-8.
- ^ Nair, Arathi; et al. (July 4, 2019). "Conceptual Evolution of Cell Signaling". International Journal of Molecular Sciences. 20 (13): 3292. doi:10.3390/ijms20133292. ISSN 1422-0067. PMC 6651758. PMID 31277491.
- ^ a b Alberts, Bruce; et al. (2002). "General Principles of Cell Communication". Molecular Biology of the Cell (4th ed.). Garland Science.
- ^ Infante, Deliana (December 2, 2024). "How Dysregulated Cell Signaling Causes Disease". News-Medical.Net. Retrieved 2025-10-05.
- ^ a b c Grosberg, R. K.; Strathmann, R. R. (2007). "The evolution of multicellularity: A minor major transition?" (PDF). Annual Review of Ecology, Evolution, and Systematics. 38: 621–654. doi:10.1146/annurev.ecolsys.36.102403.114735. Archived from the original (PDF) on 2016-03-04. Retrieved 2013-12-23.
- ^ Popper, Zoë A.; Michel, Gurvan; Hervé, Cécile; et al. (2011). "Evolution and diversity of plant cell walls: from algae to flowering plants" (PDF). Annual Review of Plant Biology. 62 (1): 567–590. Bibcode:2011AnRPB..62..567P. doi:10.1146/annurev-arplant-042110-103809. hdl:10379/6762. PMID 21351878. S2CID 11961888. Archived (PDF) from the original on 2016-07-29. Retrieved 2013-12-23.
- ^ Bonner, John Tyler (1998). "The Origins of Multicellularity" (PDF). Integrative Biology. 1 (1): 27–36. doi:10.1002/(SICI)1520-6602(1998)1:1<27::AID-INBI4>3.0.CO;2-6. ISSN 1093-4391. Archived from the original (PDF, 0.2 MB) on March 8, 2012.
- ^ Niklas, Karl J. (January 2014). "The evolutionary-developmental origins of multicellularity". American Journal of Botany. 101 (1): 6–25. Bibcode:2014AmJB..101....6N. doi:10.3732/ajb.1300314. PMID 24363320.
- ^ Albani, Abderrazak El; Bengtson, Stefan; Canfield, Donald E.; et al. (July 2010). "Large colonial organisms with coordinated growth in oxygenated environments 2.1 Gyr ago". Nature. 466 (7302): 100–104. Bibcode:2010Natur.466..100A. doi:10.1038/nature09166. PMID 20596019. S2CID 4331375.
- ^ Orgel, L. E. (December 1998). "The origin of life--a review of facts and speculations". Trends in Biochemical Sciences. 23 (12): 491–495. doi:10.1016/S0968-0004(98)01300-0. PMID 9868373.
- ^ Chatterjee, S. (2023). "The RNA World: Reality or Dogma?". From Stardust to First Cells. Springer, Cham. pp. 97–107. doi:10.1007/978-3-031-23397-5_10. ISBN 978-3-031-23397-5.
- ^ Dodd, Matthew S.; Papineau, Dominic; Grenne, Tor; et al. (1 March 2017). "Evidence for early life in Earth's oldest hydrothermal vent precipitates". Nature. 543 (7643): 60–64. Bibcode:2017Natur.543...60D. doi:10.1038/nature21377. PMID 28252057. Archived from the original on 8 September 2017. Retrieved 2 March 2017.
- ^ Betts, Holly C.; Puttick, Mark N.; Clark, James W.; Williams, Tom A.; Donoghue, Philip C. J.; Pisani, Davide (20 August 2018). "Integrated genomic and fossil evidence illuminates life's early evolution and eukaryote origin". Nature Ecology & Evolution. 2 (10): 1556–1562. Bibcode:2018NatEE...2.1556B. doi:10.1038/s41559-018-0644-x. PMC 6152910. PMID 30127539.
- ^ Griffiths, G. (December 2007). "Cell evolution and the problem of membrane topology". Nature Reviews. Molecular Cell Biology. 8 (12): 1018–1024. doi:10.1038/nrm2287. PMID 17971839. S2CID 31072778.
- ^ "First cells may have emerged because building blocks of proteins stabilized membranes". ScienceDaily. Archived from the original on 2021-09-18. Retrieved 2021-09-18.
- ^ a b c d Latorre, A.; Durban, A; Moya, A.; Pereto, J. (2011). "The role of symbiosis in eukaryotic evolution". In Gargaud, Muriel; López-Garcìa, Purificacion; Martin, H. (eds.). Origins and Evolution of Life: An astrobiological perspective. Cambridge: Cambridge University Press. pp. 326–339. ISBN 978-0-521-76131-4. Archived from the original on 24 March 2019. Retrieved 27 August 2017.
- ^ a b Weiss, Madeline C.; Sousa, F. L.; Mrnjavac, N.; et al. (2016). "The physiology and habitat of the last universal common ancestor" (PDF). Nature Microbiology. 1 (9): 16116. doi:10.1038/nmicrobiol.2016.116. PMID 27562259. S2CID 2997255.
- ^ a b Strassert, Jürgen F. H.; Irisarri, Iker; Williams, Tom A.; Burki, Fabien (25 March 2021). "A molecular timescale for eukaryote evolution with implications for the origin of red algal-derived plastids". Nature Communications. 12 (1): 1879. Bibcode:2021NatCo..12.1879S. doi:10.1038/s41467-021-22044-z. PMC 7994803. PMID 33767194.
- ^ McGrath, Casey (31 May 2022). "Highlight: Unraveling the Origins of LUCA and LECA on the Tree of Life". Genome Biology and Evolution. 14 (6) evac072. doi:10.1093/gbe/evac072. PMC 9168435.
- ^ Leander, B. S. (May 2020). "Predatory protists". Current Biology. 30 (10): R510 – R516. Bibcode:2020CBio...30.R510L. doi:10.1016/j.cub.2020.03.052. PMID 32428491. S2CID 218710816.
- ^ Gabaldón, T. (October 2021). "Origin and Early Evolution of the Eukaryotic Cell". Annual Review of Microbiology. 75 (1): 631–647. doi:10.1146/annurev-micro-090817-062213. PMID 34343017. S2CID 236916203.
- ^ Woese, C.R.; Kandler, Otto; Wheelis, Mark L. (June 1990). "Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya". Proceedings of the National Academy of Sciences of the United States of America. 87 (12): 4576–4579. Bibcode:1990PNAS...87.4576W. doi:10.1073/pnas.87.12.4576. PMC 54159. PMID 2112744.
- ^ Hooke, Robert (1665). "Observation 18". Micrographia.
- ^ Maton, Anthea (1997). Cells Building Blocks of Life. New Jersey: Prentice Hall. pp. 44-45 The Cell Theory. ISBN 978-0-13-423476-2.
- ^ a b Gest, H. (2004). "The discovery of microorganisms by Robert Hooke and Antoni Van Leeuwenhoek, fellows of the Royal Society". Notes and Records of the Royal Society of London. 58 (2): 187–201. doi:10.1098/rsnr.2004.0055. PMID 15209075. S2CID 8297229.
- ^ "History of the Cell: Discovering the Cell". education.nationalgeographic.org. Retrieved 2025-08-01.
- ^ "The Origins Of The Word 'Cell'". National Public Radio. September 17, 2010. Archived from the original on 2021-08-05. Retrieved 2021-08-05.
- ^ "cellŭla". A Latin Dictionary. Charlton T. Lewis and Charles Short. 1879. ISBN 978-1-9998557-8-9. Archived from the original on 2021-08-07. Retrieved 2021-08-05.
{{cite encyclopedia}}: CS1 maint: ignored ISBN errors (link) - ^ Hooke, Robert (1665). Micrographia: ... London: Royal Society of London. p. 113.
... I could exceedingly plainly perceive it to be all perforated and porous, much like a Honey-comb, but that the pores of it were not regular [...] these pores, or cells, [...] were indeed the first microscopical pores I ever saw, and perhaps, that were ever seen, for I had not met with any Writer or Person, that had made any mention of them before this ...
– Hooke describing his observations on a thin slice of cork. See also: Robert Hooke Archived 1997-06-06 at the Wayback Machine - ^ Schwann, Theodor (1839). Mikroskopische Untersuchungen über die Uebereinstimmung in der Struktur und dem Wachsthum der Thiere und Pflanzen. Berlin: Sander.
- ^ "4.3: Studying Cells - Cell Theory". Biology LibreTexts. 2018-07-05. Retrieved 2025-08-01.
- ^ Ribatti, Domenico (2018-03-01). "An historical note on the cell theory". Experimental Cell Research. 364 (1): 1–4. doi:10.1016/j.yexcr.2018.01.038. ISSN 0014-4827. PMID 29391153.
- ^ Ernst Ruska (January 1980). The Early Development of Electron Lenses and Electron Microscopy. Applied Optics. Vol. 25. Translated by T. Mulvey. p. 820. Bibcode:1986ApOpt..25..820R. doi:10.1364/AO.25.000820. ISBN 978-3-7776-0364-3.
- ^ Cornish-Bowden, Athel (7 December 2017). "Lynn Margulis and the origin of the eukaryotes". Journal of Theoretical Biology. The origin of mitosing cells: 50th anniversary of a classic paper by Lynn Sagan (Margulis). 434: 1. Bibcode:2017JThBi.434....1C. doi:10.1016/j.jtbi.2017.09.027. PMID 28992902.
External links
[edit]- "The Inner Life of the Cell". XVIVO website. – 2006 animation of molecular mechanisms inside cells
Cell (biology)
View on GrokipediaOverview
Definition and Key Characteristics
In biology, a cell is defined as the smallest structural and functional unit of life capable of independent reproduction and metabolism.[4] All living organisms are composed of one or more cells, which serve as the fundamental building blocks for tissues, organs, and entire multicellular bodies.[1] Cells exhibit several universal key characteristics that distinguish them as the basis of life: compartmentalization through a plasma membrane that separates the internal environment from the external one, enabling controlled exchanges; metabolism, involving enzymatic chemical reactions to process nutrients and generate energy, often stored as ATP; growth and responsiveness to environmental stimuli via receptors and signaling pathways; reproduction through cell division, ensuring propagation; and heredity encoded by DNA, which directs cellular structure, function, and inheritance.[6] These properties allow cells to maintain homeostasis, adapt to changes, and sustain life processes autonomously or in coordination within multicellular organisms.[5] Cells manifest in diverse forms, including unicellular organisms that exist as single, self-sufficient entities, such as bacteria (prokaryotes) and amoebae (eukaryotic protists), which perform all life functions independently.[7][8] In multicellular organisms like humans or plants, specialized cells collaborate, with examples including neurons for signal transmission or leaf cells for photosynthesis, yet each retains the core ability to divide and differentiate when needed.[1] Notably, viruses are distinguished from cells as non-cellular entities; they lack metabolism, independent reproduction, and cellular structures, instead relying on host cells to replicate their genetic material.[9] Cell sizes vary significantly by type, providing insight into their complexity and function. Prokaryotic cells, such as bacteria, typically range from 0.1 to 5 μm in diameter, reflecting their simpler organization. Eukaryotic cells, found in animals, plants, and fungi, are generally larger, measuring 10 to 100 μm, which accommodates membrane-bound organelles and greater internal compartmentalization. These scale differences underscore the evolutionary divergence between cell types while highlighting the cell's role as life's minimal viable unit.[5]Cell Theory
Cell theory, a cornerstone of modern biology, was formulated in the 19th century through the pioneering work of several key scientists. In 1838, German botanist Matthias Jakob Schleiden observed under the microscope that all plant tissues are aggregates of individual cells, proposing that the cell is the fundamental unit of plant structure and development; this insight was published in his seminal paper "Beiträge zur Phytogenesis" (Contributions to Phytogenesis).[10] Building on Schleiden's findings, German physiologist Theodor Schwann extended the concept to animals in 1839, demonstrating through detailed microscopic examinations that animal tissues similarly consist of cells and exhibit comparable growth patterns to plants; his comprehensive treatise, "Mikroskopische Untersuchungen über die Übereinstimmung in der Struktur und dem Wachstum der Tiere und Pflanzen" (Microscopical Researches into the Accordance in the Structure and Growth of Animals and Plants), laid the groundwork for the theory's initial principles.[11] These contributions by Schleiden and Schwann established the unifying idea that cells form the basic building blocks of all living organisms.[12] The theory was completed in 1855 by German pathologist Rudolf Virchow, who, through his studies of tissue pathology and cell division, asserted that all cells originate from pre-existing cells—a principle encapsulated in his famous axiom "omnis cellula e cellula" (every cell from a cell); this was articulated in his lectures and subsequent publication "Die Cellularpathologie in ihrer Begründung auf physiologische und pathologische Gewebelehre" (Cellular Pathology as Based Upon Physiological and Pathological Histology).[13] Virchow's addition refuted earlier notions of spontaneous generation and emphasized cellular continuity in health and disease. The resulting classical cell theory comprises three core tenets: all living organisms are composed of one or more cells; the cell is the basic unit of structure, function, and organization in organisms; and all cells arise from pre-existing cells via division. Cell theory profoundly impacts biology by providing a unified framework that connects the structure and function of life across scales, from unicellular microbes to complex multicellular organisms; it underpins disciplines such as cytology, which examines cellular structures and processes, and microbiology, which investigates cellular life at the microscopic level.[15] In the modern era, the theory has been extended to incorporate advances in genetics and biochemistry: cells carry hereditary information through DNA, which is replicated and transmitted during division; moreover, cells serve as the primary sites for metabolic activities, including energy flow via processes like cellular respiration and information processing through molecular signaling.[16] Apparent exceptions, such as syncytia—multinucleated cytoplasmic masses in structures like skeletal muscle or certain fungi—do not contradict the theory, as they form through fusion of individual cells and function as single, albeit complex, cellular units with coordinated nuclear activity.[17]History of Cell Research
Early Discoveries
The invention of the compound microscope in the late 16th and early 17th centuries, building on earlier simple lenses, enabled the first glimpses into the microscopic world, though initial instruments suffered from poor magnification and image distortion. In 1665, English scientist Robert Hooke published Micrographia, detailing his observations of thin slices of cork viewed through a microscope with up to 50x magnification; he described the honeycomb-like compartments as "cells," likening them to small rooms, and noted their porous structure composed of rigid walls. These were actually the empty lignified cell walls of dead plant tissue from cork oak (Quercus suber), marking the first documented use of the term "cell" in biology.[18][19] Concurrently in the 1660s, Italian physician Marcello Malpighi employed early microscopes to examine plant and animal tissues, revealing intricate structures such as the vascular bundles in plants and the capillary networks connecting arteries and veins in frog lungs, which he described in works like De pulmonibus (1661). His studies of insect anatomy, including the tubular structures now known as Malpighian tubules, demonstrated that tissues in both plants and animals consisted of organized, minute components, laying groundwork for comparative histology. A decade later, in the 1670s, Dutch microscopist Antonie van Leeuwenhoek crafted superior single-lens microscopes achieving 270x magnification and over 1 μm resolution; in letters to the Royal Society, he reported observing "animalcules"—tiny, motile organisms—in samples of pond water, rainwater, and dental plaque, which included protozoa like Paramecium and the first sightings of bacteria such as those in the mouth. These findings, published in Philosophical Transactions starting in 1677, expanded the known diversity of life and confirmed the ubiquity of microscopic entities.[20] Early microscopy's limitations, including chromatic and spherical aberrations that blurred images and restricted resolution to about 1-2 μm, fueled debates on cellular continuity; observers like 18th-century microscopists often misinterpreted tissue as a continuous fibrous network rather than discrete units, as finer details such as cell boundaries in living animal tissues remained elusive. In 1831, Scottish botanist Robert Brown advanced these observations by identifying a dark, opaque structure—the nucleus—within the cells of orchid (Orchidaceae) epidermal tissue during studies of fertilization, naming it in a presentation to the Linnean Society and emphasizing its consistent presence across plant cells. This discovery, detailed in his 1833 publication, highlighted internal cellular organization previously overlooked.[21][22] Building on these advances, the 1830s saw the formulation of cell theory. In 1838, German botanist Matthias Jakob Schleiden proposed that all plant tissues are composed of cells and that cells are the fundamental units of plant life. The following year, Theodor Schwann extended this idea to animals, stating that all living organisms are made up of cells. In 1855, Rudolf Virchow added the crucial insight that all cells arise from preexisting cells, completing the classical cell theory. These principles unified the understanding of life at the cellular level.[23] The transition to the 19th century brought pivotal improvements, such as achromatic lenses developed by Joseph Jackson Lister in the 1830s, which corrected color fringing and spherical distortion to achieve resolutions below 1 μm, enabling clearer views of dynamic cellular processes and paving the way for detailed investigations of cell division and morphology.[24]Development of Modern Understanding
The advent of electron microscopy in the 1930s marked a pivotal advancement in visualizing cellular ultrastructure, surpassing the resolution limits of light microscopy. Invented by Ernst Ruska and Max Knoll in 1931, the transmission electron microscope (TEM) achieved magnifications up to 100,000 times, enabling the first detailed images of subcellular components. By the 1940s, biologists like Keith Porter and Ernest Ruska applied TEM to fixed tissue sections, revealing organelles such as the endoplasmic reticulum in 1945 and mitochondria's internal cristae, which transformed understanding of cellular compartmentalization.[25][26] In 1953, James Watson and Francis Crick proposed the double-helix structure of DNA, providing a molecular framework for genetic information storage and replication within cells. This discovery, built on X-ray diffraction data from Rosalind Franklin and Maurice Wilkins, elucidated how DNA's base-pairing mechanism underpins cellular heredity and protein synthesis, bridging cytology with molecular genetics. The model implied that genetic mutations could alter cellular function, influencing subsequent research on gene expression in prokaryotes and eukaryotes.[27] Lynn Margulis advanced cellular evolution in 1967 with her endosymbiotic theory, positing that eukaryotic organelles like mitochondria and chloroplasts originated from engulfed prokaryotic symbionts. Published in the Journal of Theoretical Biology, her hypothesis integrated ultrastructural evidence from electron microscopy—such as organelles' double membranes and independent replication—with biochemical data on their prokaryote-like ribosomes and DNA. Initially controversial, the theory gained acceptance through genetic sequencing confirming shared ancestry, reshaping paradigms of cellular complexity and symbiosis.[28] The 1970s ushered in the molecular biology era with recombinant DNA technology, allowing direct manipulation of cellular genomes. Pioneered by Stanley Cohen and Herbert Boyer in 1973, they inserted foreign DNA into Escherichia coli using restriction enzymes and plasmids, creating the first recombinant organisms. This breakthrough enabled gene cloning and expression in host cells, facilitating studies of cellular processes like insulin production and accelerating biotechnology applications.[29] A major leap in cellular editing occurred in 2012 with the development of CRISPR-Cas9, a precise, programmable tool derived from bacterial immune systems. Jennifer Doudna, Emmanuelle Charpentier, and colleagues demonstrated in vitro that CRISPR-Cas9 uses guide RNA to target and cleave specific DNA sequences, enabling efficient genome modifications in eukaryotic cells. This versatility revolutionized cell engineering, from knocking out genes to study function to creating disease models. Post-2020, CRISPR advanced cellular therapies; in 2023, the FDA approved Casgevy (exagamglogene autotemcel), the first CRISPR-edited therapy for sickle cell disease, involving ex vivo editing of hematopoietic stem cells to boost fetal hemoglobin production. By 2025, over 50 CRISPR-based clinical trials targeted cellular therapies for cancers and genetic disorders, demonstrating durable engraftment and reduced off-target effects.[30][31][32] Recent decades have seen explosive growth in single-cell sequencing technologies, resolving cellular heterogeneity unattainable by bulk methods. Emerging in the early 2010s with protocols like Smart-seq (2012), the field advanced through high-throughput platforms such as Drop-seq (2015), which encapsulated cells in droplets for parallel RNA profiling of thousands. By 2025, innovations like long-read single-cell RNA sequencing integrated with spatial transcriptomics revealed dynamic gene expression in tissues, aiding discoveries in tumor microenvironments and developmental trajectories. These tools have supported thousands of studies. Optogenetics, introduced in 2005 by Karl Deisseroth and Edward Boyden, enabled light-mediated control of cellular activity via genetically encoded ion channels like channelrhodopsin-2. Expressed in target cells, these proteins allow millisecond-precision modulation of neuronal firing or other cellular signaling, transforming in vivo studies of brain circuits and muscle function. Advances through 2025 include expanded toolkits for non-neuronal cells, such as astrocytes, and two-photon excitation for deeper tissue penetration, with thousands of publications elucidating cellular networks in health and disease.[33] Synthetic biology has progressed toward engineering minimal and artificial cells, exemplified by J. Craig Venter's 2010 creation of Mycoplasma mycoides JCVI-syn1.0, the first self-replicating cell with a chemically synthesized genome. This 1.08-megabase genome, transplanted into a recipient cell, demonstrated bootable cellular life from digital code, identifying essential genes for viability. Updates by 2025 include artificial organelles, such as enzyme-loaded vesicles mimicking mitochondria for targeted ATP production, and nucleated synthetic cells with intercompartment communication via DNA nanotechnology. These constructs, often lipid-based protocells, replicate division and metabolism, paving the way for designer therapeutics. AI-assisted cell imaging has complemented these efforts; machine learning models since 2020 analyze super-resolution microscopy data to predict organelle dynamics and protein localization, enhancing synthetic design.[34][35][36]Cell Classification
Prokaryotic Cells
Prokaryotic cells are the structural and functional units of prokaryotes, which are organisms that lack a membrane-bound nucleus and other membrane-bound organelles, distinguishing them from eukaryotic cells.[5] Prokaryotes encompass the two domains of life, Bacteria and Archaea, which together represent the majority of known cellular diversity on Earth.[37] These cells typically measure between 0.1 and 5 micrometers in diameter, with archaeal cells often ranging from 0.7 to 4 micrometers.[38] Prokaryotes exhibit diverse morphologies, including cocci (spherical), bacilli (rod-shaped), and spirilla (spiral), which influence their motility and environmental interactions.[39] Prokaryotes inhabit virtually every conceivable environment, from soil and oceans to the human gut, with archaea particularly noted for thriving in extreme conditions such as hypersaline lakes, acidic hot springs, and deep-sea hydrothermal vents.[40] Their genomes are typically organized as a single circular chromosome housed in a nucleoid region, often supplemented by smaller plasmids that carry accessory genes for adaptation, such as antibiotic resistance.[41] Their diversity is vast, with over 26,000 validly described species as of November 2025, predominantly bacteria.[42]Eukaryotic Cells
Eukaryotic cells are characterized by the presence of a membrane-bound nucleus that houses their genetic material, setting them apart from prokaryotic cells, which lack this enclosure. They also feature an array of membrane-bound organelles that facilitate specialized cellular processes, contributing to their compartmentalized organization. These cells form the basis of life in the domain Eukarya, encompassing diverse kingdoms including protists, fungi, plants, and animals.[5] In terms of size and complexity, eukaryotic cells are typically much larger than prokaryotes, with diameters often ranging from 10 to 100 micrometers, which supports intricate internal structures and functions. Their genomes are substantially larger, comprising multiple linear chromosomes packaged with histone proteins within the nucleus, enabling sophisticated gene regulation. Unlike the binary fission of prokaryotes, eukaryotic cells divide through mitosis (for somatic cells) or meiosis (for gametes), precise mechanisms that distribute replicated chromosomes to daughter cells.[43][44][45] Eukaryotes display vast diversity, spanning unicellular forms such as the yeast Saccharomyces cerevisiae, a model fungus used in genetic studies, and Paramecium species, ciliated protists that exemplify free-living single-celled eukaryotes, to highly organized multicellular entities.Common Structural Features
Plasma Membrane
The plasma membrane, also known as the cell membrane, forms a selectively permeable barrier that encloses the cell and separates its internal environment from the external medium. It is primarily structured as a phospholipid bilayer, where hydrophilic heads face the aqueous environments on both sides and hydrophobic tails form the inner core, with various proteins embedded or associated within this lipid matrix. This organization is encapsulated in the fluid mosaic model, which depicts the membrane as a dynamic, two-dimensional fluid where lipids and proteins can diffuse laterally, allowing for flexibility and functionality.[46] In terms of composition, the plasma membrane typically consists of about 50% lipids and 50% proteins by weight, along with smaller amounts of carbohydrates attached to lipids (glycolipids) and proteins (glycoproteins). The lipids are predominantly phospholipids, such as phosphatidylcholine and phosphatidylethanolamine, which provide the basic bilayer framework. In eukaryotic cells, cholesterol molecules are interspersed within the bilayer, modulating membrane fluidity by restricting lipid movement at higher temperatures while preventing solidification at lower ones. Carbohydrates, often in the form of short oligosaccharide chains, contribute to cell surface diversity but comprise less than 10% of the membrane mass.[47] The plasma membrane performs essential functions that maintain cellular homeostasis and enable interaction with the environment. It exhibits selective permeability, allowing small nonpolar molecules like oxygen and carbon dioxide to pass freely via simple diffusion while restricting larger or charged substances. Transport across the membrane occurs through passive mechanisms, such as facilitated diffusion via channel proteins, and active processes, including ATP-driven pumps like the sodium-potassium ATPase, which establish ion gradients. Additionally, the membrane facilitates cell recognition through glycoproteins and glycolipids that serve as identifiers in processes like immune response, and it supports signaling by hosting receptors that detect extracellular signals and transduce them into intracellular responses, such as via G-protein coupled receptors.[47] A key aspect of membrane function involves maintaining an electrochemical gradient, which generates a membrane potential typically ranging from -20 to -90 mV in living cells. This potential arises from unequal ion distributions across the membrane, balanced by the lipid bilayer's low permeability to ions. The Nernst equation quantifies the equilibrium potential for a specific ion, below which the concentration gradient drives influx and above which electrical forces drive efflux: Here, is the gas constant, is the absolute temperature, is the ion's valence, is Faraday's constant, and and are the extracellular and intracellular ion concentrations, respectively. This equation illustrates how concentration differences create an electrochemical driving force, essential for processes like nerve impulse propagation and nutrient uptake; for example, the potassium Nernst potential in neurons is around -90 mV, contributing to the resting potential.[48] While the phospholipid-based membrane is universal, variations exist across cell types, notably in archaea, which inhabit extreme environments. Archaeal plasma membranes incorporate ether-linked lipids, where isoprenoid chains are connected via ether bonds to glycerol, unlike the ester bonds in bacterial and eukaryotic phospholipids. These ether lipids enhance stability against high temperatures, acidity, and salinity, enabling extremophile adaptations; for instance, in thermophilic archaea like Sulfolobus, they prevent lipid hydrolysis and maintain membrane integrity up to 90°C.[49]Cytoplasm and Cytoskeleton
The cytoplasm, also known as the cytosol when referring specifically to its fluid component, is the aqueous gel-like substance that fills the interior of the cell, excluding the nucleus and membrane-bound organelles. It consists primarily of water, along with dissolved ions, salts, small organic molecules, and proteins, forming a viscous medium that constitutes the majority of the cell's volume.[50][51] This gel-like consistency arises from its high concentration of macromolecules, which imparts a thickness greater than pure water, enabling dynamic changes in viscosity that facilitate intracellular motility and diffusion of materials.[50][51] The cytoplasm serves as the primary site for numerous metabolic reactions, including glycolysis and other soluble enzyme-catalyzed processes, while also acting as a storage reservoir for essential molecules such as amino acids, sugars, and ions. It provides a supportive platform for the operation and positioning of non-membrane-bound cellular components, allowing for efficient diffusion-based transport within the cell. These functions are conserved across prokaryotic and eukaryotic cells, underscoring the cytoplasm's role in maintaining cellular homeostasis and enabling rapid responses to environmental changes.[51] Embedded within the cytoplasm is the cytoskeleton, a dynamic network of protein filaments that provides structural support and enables mechanical functions in all cells. In eukaryotic cells, it comprises three main types of filaments: microtubules, assembled from α- and β-tubulin dimers with a diameter of approximately 25 nm; microfilaments, or actin filaments, formed by globular actin (G-actin) monomers polymerizing into double-helical strands about 7 nm in diameter; and intermediate filaments, diverse fibrous proteins such as keratins that assemble into rope-like structures 8–12 nm thick. Prokaryotes lack these exact structures but possess homologs, including MreB proteins that form actin-like helical filaments for shape maintenance. These components undergo continuous assembly (polymerization) and disassembly (depolymerization), driven by nucleotide hydrolysis, allowing the cytoskeleton to adapt to cellular needs. For instance, in actin treadmilling, ATP hydrolysis on incorporated G-actin subunits promotes net filament flux, with the treadmilling rate given by , where and are the association and dissociation rate constants at the filament ends.[52][53][54] The cytoskeleton plays critical roles in maintaining cell shape by resisting mechanical stresses—microtubules and microfilaments counter compression and tension, respectively, while intermediate filaments offer tensile strength—and in facilitating intracellular transport, where microtubules serve as tracks for motor proteins carrying vesicles and organelles. It is also essential for cell division, with microtubules forming the mitotic spindle to segregate chromosomes and actin filaments contracting to cleave the cytoplasm during cytokinesis. These functions highlight the cytoskeleton's integration with the cytoplasm to ensure structural integrity and dynamic processes across cell types.[52][55]Ribosomes and Protein Synthesis Machinery
Ribosomes are ribonucleoprotein complexes that serve as the primary sites for protein synthesis in all cells. In prokaryotes, the ribosome is a 70S particle composed of a small 30S subunit and a large 50S subunit; the 30S subunit contains one 16S rRNA molecule and 21 proteins, while the 50S subunit includes one 23S rRNA, one 5S rRNA, and 34 proteins.[56] In eukaryotes, the ribosome forms an 80S complex with a 40S small subunit and a 60S large subunit; the 40S subunit has one 18S rRNA and 33 proteins, whereas the 60S subunit contains one 28S rRNA (or 25S in some organisms), one 5.8S rRNA, one 5S rRNA, and 47 proteins, resulting in approximately 80 proteins total.[57] These structures enable the ribosome to decode messenger RNA (mRNA) and assemble amino acids into polypeptide chains. Ribosomes are located either free in the cytoplasm, where they synthesize soluble and nuclear proteins, or bound to the cytosolic face of the rough endoplasmic reticulum (ER) in eukaryotic cells, facilitating the co-translational insertion of proteins into the ER lumen for secretion or membrane integration.[58] In prokaryotes, ribosomes are predominantly free in the cytoplasm or associated with the plasma membrane, reflecting the absence of membrane-bound organelles.[56] The binding of ribosomes to the rough ER is mediated by signal recognition particles that recognize specific mRNA sequences, directing the machinery to appropriate locations based on protein destiny.[58] Protein synthesis on ribosomes, known as translation, proceeds in three principal stages: initiation, elongation, and termination, ensuring the accurate transfer of genetic information from mRNA to proteins. During initiation, the small ribosomal subunit binds the mRNA and the initiator tRNA, followed by large subunit joining to form the complete ribosome.[59] Elongation involves sequential addition of amino acids, while termination occurs upon recognition of stop codons, releasing the completed polypeptide.[59] The genetic code, which assigns nearly universal triplet codons on mRNA to specific amino acids via transfer RNAs (tRNAs), underpins this process across prokaryotes and eukaryotes, with minor variations in organelles or certain microbes.[60] Central to elongation is the peptidyl transferase activity within the large ribosomal subunit, where the rRNA catalyzes peptide bond formation between the growing polypeptide on the peptidyl-tRNA in the P site and the incoming aminoacyl-tRNA in the A site, achieving rates up to 20 bonds per second in bacteria.00185-2) Elongation factors, such as EF-Tu in prokaryotes and eEF1A in eukaryotes, deliver aminoacyl-tRNAs to the A site in a GTP-dependent manner, with kinetic studies showing that GTP hydrolysis accelerates tRNA accommodation and translocation, limiting overall synthesis speed to about 15-20 amino acids per second under optimal conditions.00293-1) These factors ensure fidelity and efficiency, preventing errors in codon-anticodon matching. Structural variations in ribosomes provide targets for antibiotics, such as tetracycline, which binds the 30S subunit in prokaryotes to block aminoacyl-tRNA entry into the A site, selectively inhibiting bacterial protein synthesis without affecting eukaryotic 40S subunits.[61] Recent cryo-electron microscopy (cryo-EM) structures from the 2020s have illuminated ribosome dynamics, capturing intermediate states during translocation and revealing conformational changes driven by elongation factors like EF-G, which rotate the subunits to advance mRNA and tRNAs.00908-0) These advances highlight the ribosome's mechanistic flexibility, essential for adapting to cellular demands.[62]Prokaryotic-Specific Structures
Cell Wall and Envelope
The cell wall in prokaryotes serves as a rigid protective layer external to the plasma membrane, providing structural integrity and resistance to environmental stresses. In bacteria, this wall is primarily composed of peptidoglycan, also known as murein, a complex polymer consisting of alternating units of N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM) linked by β-1,4 glycosidic bonds, with short peptide chains attached to the NAM residues that enable cross-linking.[63] This mesh-like structure forms a sacculus that encases the cell, determining its shape—such as rods, spheres, or spirals—and counteracting internal turgor pressure to prevent osmotic lysis.[63] In archaea, the cell wall analog is pseudopeptidoglycan, found in certain methanogens, which features β-1,3 glycosidic linkages between N-acetyltalosaminuronic acid and N-acetylglucosamine instead of the bacterial β-1,4 bonds, along with peptide cross-links using different amino acids like L-lysine or ornithine, conferring similar rigidity but distinct chemical properties.[64] Bacterial cell walls are classified into Gram-positive and Gram-negative types based on their response to Gram staining, reflecting differences in peptidoglycan thickness and envelope architecture. Gram-positive bacteria possess a thick peptidoglycan layer, often 20–80 nm, comprising up to 90% of the wall and interspersed with teichoic acids that anchor the wall to the membrane and contribute to ion regulation.[65] In contrast, Gram-negative bacteria have a thin peptidoglycan layer, about 2–7 nm thick, sandwiched between the inner cytoplasmic membrane and an outer membrane composed of lipopolysaccharides (LPS), which adds an additional barrier against hydrophobic compounds and host defenses.[65] The Gram-negative envelope includes a periplasmic space between the two membranes, a gel-like compartment enriched with proteins that facilitate peptidoglycan modification, nutrient degradation, and secretion of enzymes such as β-lactamases for antibiotic resistance.[65] The primary functions of the prokaryotic cell wall include maintaining cell shape under mechanical stress and providing osmotic protection, as the peptidoglycan network withstands turgor pressures up to 20 atm in some species.[63] It also plays a critical role in antibiotic susceptibility; for instance, β-lactam antibiotics like penicillin target peptidoglycan synthesis by acylating transpeptidase enzymes, preventing cross-linking and leading to cell lysis during growth.[66] Biosynthesis of peptidoglycan involves sequential steps: UDP-N-acetylglucosamine and UDP-N-acetylmuramyl-pentapeptide precursors are synthesized in the cytoplasm, lipid II carriers transport them across the membrane, and glycosyltransferases polymerize the glycan chains, followed by transpeptidation to form peptide cross-links between D-alanine and another amino acid, typically L-lysine in Gram-positives or diaminopimelic acid in Gram-negatives, creating a robust lattice.[63] While the cell wall is a defining prokaryotic feature, some eukaryotes exhibit analogous structures with different compositions. Fungal cell walls are primarily built from chitin, a β-1,4-linked polymer of N-acetylglucosamine that provides tensile strength and is cross-linked with β-glucans for rigidity.[67] In plants and algae, the wall consists mainly of cellulose, a β-1,4-linked glucose polymer forming microfibrils embedded in a matrix of hemicelluloses and pectins, enabling cell expansion and mechanical support.[68] These eukaryotic variations underscore the convergent evolution of protective barriers, though they differ fundamentally from prokaryotic peptidoglycan in biosynthesis and enzymatic vulnerabilities.Nucleoid and Genetic Organization
In prokaryotic cells, the nucleoid serves as the non-membrane-bound region housing the genomic DNA, forming an irregular complex of DNA and proteins that occupies a significant portion of the cytoplasm. This structure centers on a supercoiled circular chromosome, typically 1–4 megabases (Mb) in length, which in Escherichia coli measures approximately 4.6 Mb and adopts a compact, helical ellipsoid shape for spatial confinement within the cell.[69] The DNA's supercoiling, facilitated by topoisomerases, enables the chromosome to fit within the cell dimensions while maintaining accessibility for cellular processes.[70] The organization of the nucleoid relies on nucleoid-associated proteins (NAPs) that compact and structure the DNA into higher-order domains. Key NAPs include HU, which binds non-specifically to DNA bends and junctions to promote compaction—E. coli cells contain about 55,000 HU molecules during exponential growth—and others such as H-NS, Fis, and IHF that help form plectonemic loops, macrodomains (e.g., ~1 Mb regions like Ori and Ter in E. coli), and smaller topological domains of 10–400 kb separated by diffusion barriers.[69] Polyamines, including putrescine and spermidine, further enhance stability by neutralizing DNA's negative charge, reducing electrostatic repulsion and aiding compaction without sequence specificity.[69] This architecture contrasts sharply with the eukaryotic nucleus, which is enclosed by a double membrane that isolates chromatin from the cytoplasm, whereas the nucleoid allows immediate interaction with cytosolic components like ribosomes.[70] Functionally, the nucleoid supports coupled transcription and translation, as nascent mRNA is directly accessible to ribosomes in the cytoplasm, enabling rapid gene expression tied to the chromosome's three-dimensional organization.[69] In bacteria like Vibrio cholerae, which possess two chromosomes, each harbors a distinct replication origin (oriC I on the large chromosome and oriC II on the small one), facilitating temporally coordinated replication initiation and contributing to nucleoid dynamics.[71] During cell division, the nucleoid undergoes partitioning to ensure equal distribution to daughter cells via binary fission; the nucleoid occlusion model prevents septum formation over unsegregated DNA by inhibiting FtsZ ring assembly through proteins like SlmA in E. coli, which binds specific DNA sequences and blocks division-site maturation.[72] This dynamic reorganization, including increased long-range DNA contacts in stationary phase, maintains nucleoid integrity across growth conditions.[69]Surface Appendages
Surface appendages in prokaryotes are dynamic extracellular structures that extend from the cell envelope, enabling functions such as motility, adhesion, and environmental protection. These appendages, including flagella, pili (also known as fimbriae), and capsules, are primarily found in bacteria and archaea, where they interact with the surrounding milieu to facilitate survival and adaptation. Unlike the static cell wall, these structures are often assembled and disassembled in response to environmental cues, contributing to processes like colonization and horizontal gene transfer.[73] Bacterial flagella are long, helical filaments composed of a basal body embedded in the cell membrane and wall, a flexible hook, and an extracellular filament made primarily of flagellin proteins. The basal body acts as a rotary motor, with stator units (MotA/MotB complexes) harnessing the proton motive force across the membrane to generate torque, powering counterclockwise rotation for smooth swimming. This ion-driven mechanism, rather than ATP hydrolysis, allows flagella to achieve speeds up to 100 body lengths per second in some bacterial species, such as Vibrio alginolyticus. In archaea, the analogous structure is the archaellum, which shares a superficial rotary function but differs fundamentally in assembly: archaella are type IV pilus-like, polymerizing from the base with distinct subunits (e.g., FlaA/B) and powered by ATPases, lacking homology to bacterial flagellar components.[74] Flagella enable bacterial motility through chemotaxis, where cells exhibit a "run-and-tumble" pattern: prolonged counterclockwise rotation produces straight runs toward attractants, while clockwise switches cause filament bundling and random tumbles for reorientation. This biased random walk optimizes navigation in gradients, as seen in E. coli, where tumbling frequency decreases in favorable conditions. Pili, shorter and more numerous than flagella, include type IV variants that extend and retract via ATP-powered assembly and disassembly at the pilin tip. Retraction, driven by dedicated motors like PilT, generates forces exceeding 100 pN, facilitating twitching motility and adhesion to surfaces. Common pili (type 1 fimbriae) mediate host cell attachment in pathogens like uropathogenic E. coli, while sex pili (e.g., F-pili in conjugative plasmids) form bridges for DNA transfer during bacterial conjugation, stabilizing donor-recipient pairs under shear stress.[75][76][77] Capsules and the broader glycocalyx are polysaccharide-based layers enveloping the cell, often loosely associated with the cell wall to form a protective sheath. These structures trap water and nutrients, shielding bacteria from desiccation, phagocytosis, and antibiotics; for instance, the hyaluronic acid capsule of Streptococcus pyogenes inhibits complement activation for immune evasion. S-layers, crystalline protein arrays on many prokaryotes, further contribute to protection by masking surface antigens and promoting biofilm formation, where appendages like pili and capsules anchor communities. In biofilms, glycocalyx matrices enhance adhesion and resistance, as exemplified by Pseudomonas aeruginosa exopolysaccharides that withstand host defenses.[78] Recent advances in biofilm engineering leverage these appendages for biotechnological applications, such as designing synthetic microbial consortia for wastewater remediation and medical implants. For example, engineered E. coli capsules have been modified since 2020 to form tunable biofilms that deliver therapeutics in the gut, improving probiotic efficacy and reducing pathogen colonization. As of 2025, engineered probiotics continue to advance, with studies demonstrating precise modulation of the intestinal microenvironment for treating inflammatory bowel disease through targeted payload delivery.[79][80]Eukaryotic-Specific Structures
Nucleus
The nucleus serves as the control center of eukaryotic cells, housing the genetic material and orchestrating key processes such as gene expression and DNA replication. Enclosed by the nuclear envelope, it separates the genomic DNA from the cytoplasm, enabling precise regulation of transcription and RNA processing that is characteristic of eukaryotic complexity. This organelle is essential for maintaining cellular identity and responding to environmental cues through controlled gene activity. The nuclear envelope consists of a double lipid bilayer—the inner and outer nuclear membranes—perforated by nuclear pore complexes (NPCs) that facilitate selective transport between the nucleus and cytoplasm. The inner nuclear membrane is lined by the nuclear lamina, a meshwork of type V intermediate filaments primarily composed of lamin proteins, which provides structural support and anchors chromatin to the envelope. Chromatin, the complex of DNA and histone proteins, is organized into euchromatin, which is loosely packed and transcriptionally active, and heterochromatin, which is densely condensed and generally inactive; this packaging modulates access to genetic information. Within the nucleus, the nucleolus forms a prominent substructure dedicated to ribosomal RNA (rRNA) synthesis and ribosome assembly, containing high concentrations of RNA polymerase I and ribosomal proteins. The primary functions of the nucleus include the storage and protection of DNA, as well as the initiation of transcription where DNA is copied into messenger RNA (mRNA) by RNA polymerases. This process underlies the central dogma of molecular biology, in which genetic information flows from DNA to RNA and subsequently to proteins, with transcription occurring exclusively in the nucleus. Mature mRNA is then exported to the cytoplasm through NPCs via the Ran GTPase cycle, a GTP-binding protein system that generates a nuclear-cytoplasmic gradient to drive directional transport; Ran-GTP promotes export complex assembly in the nucleus, while its hydrolysis in the cytoplasm disassembles them. The nucleolus supports this by producing rRNA components essential for translation machinery. During the cell cycle, the nucleus exhibits dynamic changes, particularly in mitosis, where the nuclear envelope breaks down to allow chromosome segregation. Phosphorylation of lamin proteins by cyclin-dependent kinases causes the lamina to depolymerize into dimers, leading to envelope disassembly and chromatin condensation. Post-mitosis, dephosphorylation enables lamina reassembly and envelope reformation around daughter nuclei. These dynamics ensure equitable distribution of genetic material while temporarily suspending transcription. Variations in nuclear organization occur in certain cell types, such as multinucleated syncytia like skeletal muscle fibers, where hundreds of nuclei share a common cytoplasm to support high metabolic demands and rapid protein synthesis. In these cells, nuclei maintain individual identities but coordinate gene expression to optimize tissue function, illustrating adaptations in nuclear architecture for specialized roles.Membrane-Bound Organelles
Membrane-bound organelles in eukaryotic cells enable compartmentalization, allowing specialized biochemical reactions to occur in isolated environments while facilitating communication through vesicular transport. These include the endomembrane system components such as the endoplasmic reticulum, Golgi apparatus, and lysosomes, as well as other organelles like peroxisomes, mitochondria, and chloroplasts (in photosynthetic eukaryotes), each with distinct roles in protein processing, lipid metabolism, degradation, detoxification, energy production, and photosynthesis.[81] The endoplasmic reticulum (ER) is a continuous network of tubules and sacs that spans the cytoplasm, divided into rough and smooth domains based on ribosomal association and function. The rough ER, studded with ribosomes on its cytoplasmic surface, serves as the primary site for co-translational folding and modification of secretory and membrane proteins, where chaperones like BiP assist in attaining native conformations to prevent aggregation.[82] In contrast, the smooth ER lacks ribosomes and specializes in lipid synthesis, including phospholipids and steroids essential for membrane biogenesis, as well as calcium ion storage in its lumen via pumps like SERCA, which buffers cytosolic Ca²⁺ levels for signaling.[81][83] The Golgi apparatus consists of stacked cisternae organized into cis, medial, and trans compartments, exhibiting polarity with the cis face receiving vesicles from the ER and the trans face directing cargo to destinations like the plasma membrane or lysosomes. It functions in post-translational modification, particularly glycosylation, where N-linked and O-linked sugars are added to proteins by resident glycosyltransferases, influencing protein stability, trafficking, and cell-cell recognition.[84] Sorting occurs here via mannose-6-phosphate tags for lysosomal enzymes or clathrin adaptors for endocytic pathways, ensuring precise delivery. Lysosomes are acidic vesicles (pH ≈5) containing over 50 hydrolytic enzymes, including acid hydrolases like cathepsins and nucleases, that degrade macromolecules from endocytosis, phagocytosis, or autophagy.[85] The low pH, maintained by V-ATPase proton pumps, optimizes hydrolase activity for breaking down proteins, lipids, and nucleic acids into reusable monomers.[86] In autophagy, lysosomes fuse with autophagosomes to digest damaged organelles or cytosolic components, recycling nutrients during stress.[87] Plant vacuoles serve analogous roles, combining degradative functions with turgor maintenance. Mitochondria are double-membrane-bound organelles present in nearly all eukaryotic cells, serving as the primary sites of aerobic respiration and ATP production via oxidative phosphorylation. The outer membrane is freely permeable to small molecules, while the highly folded inner membrane forms cristae that house the electron transport chain complexes and ATP synthase. Mitochondria possess their own circular genome (mtDNA) and ribosomes, enabling semi-autonomous protein synthesis, and originated from endosymbiotic alpha-proteobacteria. Beyond energy production, they regulate cellular metabolism, calcium homeostasis, apoptosis through cytochrome c release, and generate reactive oxygen species (ROS) as signaling molecules. Dysfunctional mitochondria are implicated in aging and diseases like Parkinson's.[88] Chloroplasts are specialized membrane-bound organelles found in the cells of plants and algae, responsible for photosynthesis, which converts light energy into chemical energy stored in glucose. Enclosed by a double membrane, they contain internal thylakoid membranes stacked into grana, where photosystems I and II capture light to drive electron transport and generate ATP and NADPH. The fluid stroma surrounding the thylakoids hosts the Calvin-Benson cycle for carbon fixation, along with chloroplast DNA (cpDNA) and ribosomes, reflecting their endosymbiotic origin from cyanobacteria. Chloroplasts also synthesize fatty acids, amino acids, and hormones, and coordinate with the nucleus via retrograde signaling.[89] Peroxisomes are single-membrane organelles that perform β-oxidation of very long-chain fatty acids, shortening them for mitochondrial processing, and detoxify reactive oxygen species (ROS) like hydrogen peroxide using catalase and peroxidases. This dual role positions peroxisomes as key regulators of lipid homeostasis and oxidative balance, with impaired function linked to disorders like Zellweger syndrome. Recent studies highlight peroxisome-mitochondria crosstalk at contact sites, where shared metabolites influence ROS signaling in aging.[90] The endomembrane system organelles integrate via vesicular transport, where COPII coats mediate anterograde ER-to-Golgi trafficking, COPI handles retrograde Golgi-to-ER recycling, and clathrin coats facilitate sorting from the trans-Golgi to lysosomes or the plasma membrane.[91] This dynamic network maintains cellular homeostasis by coordinating synthesis, modification, and degradation.Extracellular Matrix
The extracellular matrix (ECM) in eukaryotic cells is a complex network of secreted macromolecules that surrounds and supports cells in tissues, providing both structural integrity and biochemical cues essential for cellular behavior. Primarily found in animal tissues, the ECM differs fundamentally from the rigid cellulose-based cell walls of plants, which offer mechanical protection but lack the dynamic flexibility and signaling capabilities of the animal ECM. In animals, the ECM enables tissue resilience and adaptability, allowing for processes like wound healing and development.[92] The composition of the ECM includes fibrous proteins such as collagens, which form the structural scaffold; elastin, which imparts elasticity; and glycoproteins like fibronectin, which facilitate cell adhesion. Additionally, proteoglycans—core proteins decorated with long glycosaminoglycan (GAG) chains—contribute to the hydrated ground substance that fills the matrix, providing a gel-like medium for diffusion and lubrication. Collagens, the most abundant proteins, assemble into fibrils that vary by type (e.g., type I in tendons for tensile strength), while GAGs like hyaluronic acid attract water to maintain tissue hydration and compressive resistance.[92][93][94] The ECM serves multiple functions, including mechanical support to withstand physiological stresses, mediation of cell adhesion through integrins that link the matrix to the cytoskeleton, and regulation of signaling pathways via sequestered growth factors such as transforming growth factor-beta (TGF-β). Integrins, transmembrane receptors, bind ECM components like fibronectin and collagen, transmitting signals that influence cell proliferation, migration, and survival. Growth factors embedded in the matrix are released upon remodeling, modulating cellular responses in development and repair.[93][92][94] ECM types include the basal lamina, a thin (approximately 50-100 nm) sheet-like structure underlying epithelial and endothelial cells, composed mainly of type IV collagen, laminin, and proteoglycans to provide a barrier and filtration function. In contrast, the interstitial matrix is a looser, fibrillar network in connective tissues, rich in type I and III collagens and elastin, filling spaces between cells to enable flexibility and nutrient exchange. These distinct architectures support specialized tissue roles, such as the basal lamina's role in organ compartmentalization.[95][93] ECM dynamics involve continuous remodeling, primarily driven by matrix metalloproteinases (MMPs), a family of enzymes that degrade ECM components to facilitate tissue morphogenesis, angiogenesis, and repair. Dysregulated MMP activity can lead to excessive matrix deposition and fibrosis, a pathological scarring process seen in diseases like liver cirrhosis, where persistent TGF-β signaling promotes fibroblast activation and collagen overproduction. This remodeling balance is crucial for maintaining tissue homeostasis.[96][97]Cellular Processes
Metabolism and Energy Production
Cellular metabolism consists of catabolic pathways that degrade complex molecules to release energy and anabolic pathways that utilize this energy to synthesize cellular components. Catabolism provides the free energy needed for anabolism and other energy-demanding processes, while anabolism builds essential biomolecules such as proteins, nucleic acids, and lipids. Central to these processes is adenosine triphosphate (ATP), the primary energy currency of the cell, which stores and transfers energy through the hydrolysis of its phosphoanhydride bonds, releasing approximately 30.5 kJ/mol under standard conditions.[98][99][100] Glycolysis represents the foundational catabolic pathway for glucose breakdown, occurring in the cytosol of both prokaryotic and eukaryotic cells and comprising ten sequential enzymatic reactions divided into an energy-investment phase and an energy-payoff phase. In the investment phase, two ATP molecules are consumed to phosphorylate glucose and its derivatives, while the payoff phase generates four ATP through substrate-level phosphorylation and two NADH by oxidizing glyceraldehyde-3-phosphate. The net yield per glucose molecule is two ATP and two NADH, with the overall reaction summarized as: This pathway is universal and evolutionarily ancient, enabling rapid ATP production even in the absence of oxygen.[101][102][103] Under aerobic conditions, pyruvate from glycolysis is transported into mitochondria (in eukaryotes) or the cytoplasm (in prokaryotes with analogous systems), where it is decarboxylated to acetyl-CoA and enters the citric acid cycle, producing additional NADH and FADH₂. These electron carriers donate electrons to the electron transport chain (ETC), a series of four transmembrane protein complexes (I–IV) embedded in the inner mitochondrial membrane or plasma membrane. Complex I (NADH dehydrogenase) and complex II (succinate dehydrogenase) feed electrons from NADH and FADH₂, respectively, which are then transferred through complexes III (cytochrome bc₁) and IV (cytochrome c oxidase) to oxygen, the terminal acceptor, forming water. This electron flow drives proton pumping across the membrane, establishing an electrochemical gradient (proton-motive force). ATP synthesis occurs via chemiosmosis, where protons flow back through ATP synthase (complex V), rotating its rotor to catalyze ADP phosphorylation, yielding up to 30–32 ATP per glucose molecule when coupled with glycolysis and the citric acid cycle.[104][105][106] In anaerobic environments, common among prokaryotes, oxidative phosphorylation is unavailable, so cells rely on fermentation to regenerate NAD⁺ for continued glycolysis. Fermentation pathways, such as lactic acid fermentation in some bacteria or alcoholic fermentation in yeast, convert pyruvate to byproducts like lactate or ethanol, yielding no net ATP beyond the two from glycolysis but allowing ATP production at rates up to 100 times faster than aerobic respiration under low-oxygen conditions. In photosynthetic organisms like plant cells, chloroplasts perform light-dependent reactions that generate ATP and NADPH via a proton gradient across the thylakoid membrane, powering carbon fixation in the [Calvin cycle](/page/Calvin cycle) to produce glucose as an energy storage form.[107][108][109] Metabolic flux through these pathways is precisely regulated to balance energy supply and demand, primarily via allosteric modulation of enzymes. For example, phosphofructokinase-1 (PFK-1), which catalyzes the committed step of glycolysis (fructose-6-phosphate to fructose-1,6-bisphosphate), is allosterically activated by AMP and fructose-2,6-bisphosphate when energy is low, but inhibited by high ATP and citrate levels to prevent unnecessary glucose consumption. Similar feedback mechanisms control the citric acid cycle and ETC, ensuring efficient resource allocation; for instance, ATP utilization in protein synthesis on ribosomes consumes a significant portion of cellular ATP, linking metabolism to biosynthetic demands.[110][111][112]Growth, Division, and Reproduction
Cell growth refers to the increase in cellular mass and volume, primarily occurring during the interphase of the cell cycle, which consists of three main phases: G1, S, and G2. In the G1 phase, the cell synthesizes proteins and organelles to prepare for DNA replication, ensuring sufficient resources for subsequent division. The S phase involves DNA synthesis, where the genetic material is duplicated to produce identical sister chromatids. The G2 phase allows for further growth and checks for DNA replication errors before entering mitosis. These phases are tightly regulated by checkpoints to prevent errors; for instance, the G1/S checkpoint assesses DNA damage, with the tumor suppressor protein p53 activating repair mechanisms or halting progression if damage is irreparable.[113][114][115] Prokaryotic cells primarily reproduce through binary fission, an asexual process where the cell elongates, replicates its circular chromosome, and divides into two genetically identical daughter cells. Central to this mechanism is the Z-ring, formed by polymerization of the tubulin-like GTPase protein FtsZ, which assembles at the midcell division site and constricts the membrane inward, guiding septum formation. FtsZ's GTPase activity drives treadmilling of protofilaments, enabling dynamic ring contraction and coordination with other divisome proteins for efficient cytokinesis. This process typically takes 20-60 minutes in bacteria like Escherichia coli, allowing rapid population growth under favorable conditions.[116][117][118] In eukaryotes, cell division occurs via mitosis for somatic cells or meiosis for gametes, both preceded by interphase growth. Mitosis ensures equal distribution of replicated chromosomes to two daughter cells and comprises four phases: prophase, where chromosomes condense and the mitotic spindle—a microtubule-based apparatus—begins forming from centrosomes; metaphase, in which chromosomes align at the equatorial plate via kinetochore attachments to spindle fibers; anaphase, where sister chromatids separate and migrate to opposite poles driven by microtubule shortening; and telophase, marked by nuclear envelope reformation and decondensation of chromosomes. Cytokinesis follows, dividing the cytoplasm via an actin-myosin contractile ring in animal cells or a cell plate in plants. Meiosis, essential for sexual reproduction, involves two divisions: meiosis I reduces chromosome number by separating homologous pairs (with crossing over in prophase I for genetic diversity), followed by meiosis II, which resembles mitosis to yield four haploid cells.[115][119][120] Cell cycle progression is regulated by cyclin-dependent kinases (CDKs), serine/threonine kinases activated by binding to phase-specific cyclins, which fluctuate in concentration to drive transitions—such as cyclin D-CDK4/6 for G1, cyclin E-CDK2 for G1/S, cyclin A-CDK2 for S/G2, and cyclin B-CDK1 for G2/M. Checkpoints monitor fidelity; dysregulation, like unchecked proliferation, can trigger apoptosis, a programmed cell death pathway involving caspase activation to eliminate damaged cells and maintain tissue homeostasis. For example, p53 induces apoptosis via Bax/Bak if DNA damage persists beyond repair thresholds.[121][122][114] At the cellular level, asexual reproduction predominates in prokaryotes via binary fission and in unicellular eukaryotes via mitosis, producing clonal offspring for efficient propagation in stable environments. Sexual reproduction, conversely, relies on meiosis to generate haploid gametes that fuse during fertilization, introducing genetic variation through recombination and independent assortment, which enhances adaptability in eukaryotes. This distinction underscores mitosis and binary fission's role in growth and maintenance versus meiosis's contribution to diversity.[118][119][123]DNA Replication and Repair
DNA replication is a fundamental process in which genetic information is duplicated to ensure accurate transmission to daughter cells during cell division. In both prokaryotes and eukaryotes, replication proceeds semi-conservatively, meaning each new DNA double helix consists of one parental strand and one newly synthesized strand, as demonstrated by the classic density-labeling experiments using isotopes of nitrogen in Escherichia coli. This mechanism was confirmed through centrifugation analysis showing hybrid density DNA after one generation and a mix of hybrid and light DNA after subsequent generations. Replication initiates at specific origins of replication, where helicase unwinds the double helix, forming a replication fork that moves bidirectionally. In prokaryotes, such as E. coli, the primary replicative polymerase is DNA polymerase III (Pol III), a holoenzyme complex with high processivity that synthesizes both leading and lagging strands. Pol III incorporates nucleotides at a rate of approximately 1000 nucleotides per second per fork, enabling rapid genome duplication. In eukaryotes, replication is more complex due to larger genomes and multiple origins; DNA polymerase δ (Pol δ) primarily synthesizes the lagging strand, while DNA polymerase ε (Pol ε) handles the leading strand, both operating at about 50 nucleotides per second. The lagging strand is synthesized discontinuously as short Okazaki fragments, each primed by RNA primase and later joined by DNA ligase after primer removal; this was first observed in E. coli studies revealing nascent DNA segments of 1000–2000 nucleotides. Proofreading by the 3'→5' exonuclease activity of these polymerases reduces incorporation errors to approximately per nucleotide, enhancing fidelity. Despite these safeguards, DNA is susceptible to damage from endogenous and environmental sources, necessitating repair mechanisms to maintain genomic integrity. Base excision repair (BER) addresses small, non-helix-distorting lesions like oxidative damage or deamination; DNA glycosylases recognize and excise the damaged base, creating an apurinic/apyrimidinic (AP) site that is processed by AP endonuclease and repaired using the intact strand as a template. Nucleotide excision repair (NER) removes bulky, helix-distorting adducts, such as those induced by ultraviolet (UV) light forming cyclobutane pyrimidine dimers; in eukaryotes, this involves damage recognition by XPC or RNA polymerase stalling, excision of a 24–32 nucleotide oligonucleotide by XPF-ERCC1 and XPG, and resynthesis. Mismatch repair (MMR) corrects base-pairing errors or small insertion/deletion loops from replication slippage; MutS homologs (MSH proteins, e.g., MSH2-MSH6) recognize mismatches, followed by excision and resynthesis directed by strand discrimination. Double-strand breaks (DSBs), the most severe lesions, are repaired by homologous recombination (HR) or non-homologous end joining (NHEJ). HR, predominant in S/G2 phases, uses a sister chromatid template for error-free repair via strand invasion and synthesis, as elucidated in early yeast models. NHEJ ligates broken ends directly, often introducing small insertions/deletions, and is active throughout the cell cycle via Ku proteins and DNA-PKcs. Telomeres, the repetitive ends of linear eukaryotic chromosomes, pose a replication challenge due to the end-replication problem; telomerase, a reverse transcriptase with an integral RNA component serving as a template (e.g., 3'-CAAUCCCAAUC-5' in humans), extends 3' overhangs by adding TTAGGG repeats, preventing progressive shortening and senescence. Errors persisting through replication and repair can lead to mutations, including point mutations, insertions, deletions, or chromosomal rearrangements, which accumulate and drive oncogenesis. Defects in replication fidelity or repair pathways, such as polymerase proofreading deficiencies or MMR inactivation, elevate mutation rates and are implicated in ~10–15% of cancers, including colorectal tumors with microsatellite instability.Motility and Movement
Cells exhibit motility through diverse mechanisms that enable locomotion across surfaces or through fluids, essential for processes such as nutrient acquisition, predator avoidance, and colonization. In prokaryotes, flagellar rotation drives swimming motility via a rotary motor embedded in the cell membrane, powered by proton motive force, achieving rotation speeds of 100-300 Hz under optimal conditions. This helical flagellar filament propels bacteria like Escherichia coli at velocities up to 20-30 body lengths per second. Gliding motility, observed in bacteria such as Myxococcus xanthus, relies on type IV pili, which extend and retract to pull the cell forward across solid surfaces at speeds of 2-5 μm/min, without requiring flagella. Eukaryotic cells employ more varied strategies for extracellular movement. Cilia and flagella feature a characteristic 9+2 arrangement of microtubules, where outer dynein arms generate ATP-powered sliding between doublets, resulting in bending waves that propel cells like Chlamydomonas at 100-200 μm/s. Amoeboid locomotion, typical of leukocytes and amoebae, involves actin polymerization at the leading edge to form pseudopodia, enabling irregular crawling through tissues at rates of 0.1-1 μm/min. Similarly, many adherent cells crawl using lamellipodia, broad actin-rich protrusions driven by Arp2/3-mediated branching polymerization, facilitating migration at 0.5-10 μm/min on substrates. Intracellular motility transports vesicles and organelles along cytoskeletal tracks, primarily microtubules, using motor proteins kinesin and dynein. Kinesin moves toward the microtubule plus end with an 8 nm step size per ATP hydrolyzed, achieving velocities of 0.2-0.8 μm/s, while dynein travels to the minus end with variable steps of 8-32 nm and speeds up to 1 μm/s. The velocity of these motors is given by , where is the step size and is the dwell time between steps. All these mechanisms are fueled by ATP hydrolysis, which provides the chemical energy for conformational changes in motor proteins. Cellular motility often responds to environmental cues, such as phototaxis in photosynthetic protists like Euglena, where light gradients direct flagellar beating toward optimal illumination, and geotaxis (or gravitaxis) in dinoflagellates, enabling upward or downward migration in response to gravity via reorientation of swimming direction. Emerging biohybrid nanomotors, integrating synthetic components with cellular elements, show promise for enhanced motility control, though applications remain in early development as of 2025.Cell Communication and Signaling
Intracellular Signaling
Intracellular signaling, also known as signal transduction, refers to the processes by which cells convert external or internal stimuli into specific cellular responses through a series of molecular interactions within the cytoplasm and nucleus. These pathways enable cells to sense changes in their environment and regulate functions such as metabolism, growth, and survival. Key components include receptors that detect signals, second messengers that amplify them, and kinase cascades that propagate the response, often culminating in transcriptional changes. Dysregulation of these pathways is implicated in diseases like cancer.00665-3) Receptors such as G-protein-coupled receptors (GPCRs), which feature seven transmembrane domains, initiate signaling by activating heterotrimeric G proteins upon ligand binding, leading to the dissociation of Gα and Gβγ subunits that modulate downstream effectors. Enzyme-linked receptors, particularly receptor tyrosine kinases (RTKs), dimerize and autophosphorylate upon activation, creating docking sites for adaptor proteins and initiating phosphorylation relays. These receptor types transduce signals through distinct but sometimes overlapping intracellular mechanisms.[124]00665-3) Second messengers play a central role in amplifying signals. Cyclic adenosine monophosphate (cAMP), produced by adenylyl cyclase upon GPCR activation, activates protein kinase A (PKA), which phosphorylates targets to influence gene expression and metabolism. Inositol 1,4,5-trisphosphate (IP3) and calcium ions (Ca²⁺) are released from the endoplasmic reticulum following phospholipase C activation, where IP3 binds IP3 receptors to trigger Ca²⁺ efflux, forming oscillatory waves that regulate processes like muscle contraction and secretion. These messengers enable rapid, diffusible propagation of signals within the cell.[125][126] Cascade pathways, such as the mitogen-activated protein kinase (MAPK) pathway, involve sequential phosphorylation events. In the ERK branch, receptor activation leads to Ras GTP loading, which recruits Raf kinase; Raf then phosphorylates and activates MEK, which in turn phosphorylates ERK, promoting translocation to the nucleus for transcription factor activation and cell proliferation. This modular cascade allows integration of multiple inputs and fine-tuned outputs.[127] A prominent example is insulin signaling, where the insulin receptor, an RTK, phosphorylates insulin receptor substrates (IRS), recruiting phosphatidylinositol 3-kinase (PI3K) to generate PIP3, which activates Akt kinase. Akt promotes glucose uptake by inhibiting GSK3 and translocating GLUT4 transporters to the membrane, ensuring metabolic homeostasis. This pathway exemplifies how signaling integrates nutrient sensing with cellular responses.[128] Negative feedback mechanisms maintain signaling fidelity and prevent overstimulation. Phosphatases, such as protein tyrosine phosphatases (PTPs), dephosphorylate activated kinases in RTK and MAPK pathways, terminating the signal; for instance, MAPK phosphatases (MKPs) specifically inactivate ERK to reset the cascade. These loops ensure transient, proportional responses to stimuli.30197-0) Dysregulation of intracellular signaling often drives oncogenesis, with mutations in oncogenes like RAS leading to constitutive MAPK activation and uncontrolled proliferation in cancers such as pancreatic adenocarcinoma. Similarly, PI3K/Akt hyperactivation from PTEN loss promotes survival and metastasis. Targeted therapies, like MEK inhibitors, exploit these vulnerabilities.[127][128]Intercellular Communication
Intercellular communication enables cells within multicellular organisms to coordinate behaviors essential for development, homeostasis, and response to environmental cues. This process involves the transmission of signals between cells, often through specific molecules that bind to receptors on target cells, triggering responses that maintain tissue integrity and organismal function. In contrast to intracellular signaling, which handles internal processing, intercellular signaling focuses on external exchanges that integrate collective cellular activities.[129] Cells employ diverse mechanisms for intercellular communication, categorized by the distance and mode of signal transmission. Autocrine signaling occurs when a cell releases a ligand that binds to receptors on its own surface, influencing its own activity; for example, immune cells like T lymphocytes use autocrine signaling with interleukins to amplify their responses. Direct communication occurs via physical connections, such as gap junctions in animal cells, which form channels composed of connexin proteins allowing the passage of ions, second messengers, and small metabolites between adjacent cytoplasms. In plants, plasmodesmata serve a similar role, creating symplastic pathways through cell walls for the transport of nutrients and signaling molecules like sugars and hormones. Paracrine signaling involves local diffusion of signaling molecules to nearby cells; for instance, histamine released by mast cells induces bronchoconstriction and inflammation in adjacent bronchial smooth muscle cells via H1 receptors. Endocrine signaling transmits hormones, such as insulin, through the bloodstream to distant target cells, regulating systemic processes like glucose metabolism by binding to specific receptors with high affinity. Synaptic signaling, a specialized paracrine form, occurs at neuronal junctions where neurotransmitters like acetylcholine are released into the synaptic cleft to rapidly propagate electrical impulses across short distances.[130][131][132][129][129][129][129] Key signaling molecules include cytokines and growth factors, which mediate ligand-receptor interactions with precise binding affinities. Cytokines, such as interleukins, act as paracrine or endocrine signals to orchestrate immune responses; excessive release can lead to cytokine storms, where hyperactivation of immune cells causes widespread inflammation and tissue damage, as observed in severe infections like COVID-19. Growth factors like epidermal growth factor (EGF) bind to the EGF receptor (EGFR) with a dissociation constant (Kd) of approximately 0.1-1 nM, promoting cell proliferation and differentiation through dimerization and autophosphorylation of the receptor. In development, the Notch-Delta pathway exemplifies juxtacrine signaling, where membrane-bound Delta ligands on one cell interact directly with Notch receptors on adjacent cells, cleaving the intracellular Notch domain to regulate cell fate decisions, such as neurogenesis and somitogenesis.[129][133][134][135] The evolutionary origins of intercellular communication trace back to unicellular organisms, where bacterial quorum sensing using autoinducer molecules like acyl-homoserine lactones enables population-level coordination of behaviors such as biofilm formation. This primitive signaling evolved into sophisticated multicellular systems, adapting mechanisms like diffusible signals into contact-dependent pathways to support tissue organization in eukaryotes. Recent advances highlight exosome-mediated communication, where extracellular vesicles carrying proteins, lipids, and RNAs transfer information between cells, contributing to disease progression; for example, tumor-derived exosomes promote metastasis by altering stromal cell behavior in cancer, and dysregulated exosomal cargo exacerbates neuroinflammation in neurodegenerative disorders like Alzheimer's disease.[136][137][138]Multicellularity
Cell Differentiation and Specialization
Cell differentiation is the process by which unspecialized cells in multicellular organisms acquire distinct structures and functions, enabling the formation of diverse tissues and organs essential for complex body plans.00282-7) In animals, this begins with totipotent zygotes that can give rise to all cell types, including extraembryonic tissues; these transition to pluripotent embryonic stem cells capable of forming any body cell lineage, followed by multipotent stem cells restricted to specific tissues, and ultimately terminally differentiated cells like neurons.00039-0) For instance, neural stem cells differentiate into mature neurons through sequential stages involving progenitor proliferation and lineage commitment, driven by environmental cues and intrinsic factors.[139] Epigenetic modifications play a central role in locking in these differentiated states by altering gene accessibility without changing the DNA sequence. DNA methylation typically represses gene expression by adding methyl groups to cytosine bases in promoter regions, silencing pluripotency genes as cells specialize, while histone modifications—such as acetylation promoting open chromatin for active transcription or methylation variants enabling repression—fine-tune lineage-specific programs.[140] These changes ensure stable inheritance of cellular identity during division, preventing reversion to less committed states under normal conditions.[141] Gene regulation during differentiation is orchestrated by master transcription factors that activate or repress cascades of target genes. Hox genes, a family of homeobox-containing transcription factors, pattern the anterior-posterior axis in animal embryos by specifying segmental identities; their clustered genomic organization and collinear expression along the body axis, first elucidated in Drosophila, ensure precise spatial control of development.[142] Similarly, tissue-specific factors like MyoD, a basic helix-loop-helix protein discovered in 1987, drive skeletal muscle differentiation by binding enhancers to induce myogenic genes, converting non-muscle cells into myoblasts and myotubes.[143] A prominent example of differentiation is hematopoiesis, where hematopoietic stem cells in the bone marrow give rise to all blood cell types through a hierarchical process: multipotent progenitors commit to myeloid or lymphoid lineages, yielding erythrocytes, platelets, and various leukocytes, regulated by cytokines and transcription factors to meet physiological demands.00125-6) In plants, totipotency is more widespread, allowing differentiated somatic cells—such as leaf protoplasts—to dedifferentiate and regenerate entire plants via somatic embryogenesis, a plasticity exploited in tissue culture for propagation.[144] Cellular plasticity challenges the irreversibility of differentiation, as demonstrated by induced pluripotent stem cells (iPSCs). In a seminal 2006 study, introduction of four transcription factors—Oct4, Sox2, Klf4, and c-Myc (Yamanaka factors)—reprogrammed mouse fibroblasts to a pluripotent state resembling embryonic stem cells, enabling differentiation into any cell type and opening avenues for regenerative medicine.00976-7) Conversely, pathological dedifferentiation occurs in cancer, where tumor cells revert to a stem-like state, reacquiring proliferative and migratory capacities; this process, often triggered by epigenetic dysregulation or signaling aberrations, underlies tumor heterogeneity and resistance to therapy.[145]Evolution of Multicellular Organisms
The transition from unicellular to multicellular life represents a major evolutionary milestone, with evidence indicating that multicellular eukaryotes emerged around 1.6 billion years ago. Fossil discoveries from formations in China, India, and Australia reveal microscopic, algal-like organisms such as Qingshania magnifica, consisting of strings of up to 20 cells with distinct walls and spores, pushing back the timeline of multicellularity by nearly a billion years from previous estimates.[146] These early forms likely arose from eukaryotic ancestors that had already developed complex body plans, including cylindrical and spherical structures, as seen in 1.642-billion-year-old fossils from Australia.[147] Morphological and chemical analyses of 1.6-billion-year-old fossils from India further support this timeline by revealing preserved cellular structures and eukaryotic-specific chemical signatures that distinguish these multicellular clusters from prokaryotic mats.[148] Mechanisms facilitating this transition involved the evolution of cell adhesion and cooperation, progressing from loose colonial aggregates to integrated multicellular organisms with division of labor. In animals, cadherins—calcium-dependent adhesion proteins—played a pivotal role, with classical cadherins emerging in the metazoan lineage to form adherens junctions via binding to β-catenin, enabling stable cell-cell contacts essential for tissue formation.[149] Premetazoan precursors like choanoflagellates, the closest unicellular relatives to animals, exhibit cadherin-like proteins and form multicellular colonies through direct microvilli contacts and actomyosin-mediated contractility, foreshadowing animal morphogenesis.[150] Similarly, in the green algal lineage leading to plants, volvocine algae such as Volvox evolved multicellularity through extracellular matrix embedding and coordinated cell division, with genomic evidence showing co-option of cell cycle regulators for specialization around 234 million years ago during the Triassic.[151] This shift from colonial to true multicellularity imposed a germline-soma distinction, where somatic cells support reproduction by germline cells, resolving intracellular conflicts but requiring mechanisms to suppress selfish mutations.[152] Multicellularity conferred significant advantages, including increased body size for enhanced resource acquisition and resistance to predation, while allowing cellular specialization for efficient division of labor. Experimental evolution in unicellular algae like Chlamydomonas reinhardtii demonstrates that multicellular clusters reduce predation rates by up to 2.5-fold against predators such as rotifers, as larger sizes exceed filtration thresholds.[153] In animals, this enabled the development of specialized functions, such as nutrient uptake in choanoflagellate-like collar cells, paving the way for complex tissues. However, costs include elevated bioenergetic demands for maintaining intercellular connections and the evolutionary trade-off of reduced individual motility, as seen in volvocine algae where multicellular forms sacrifice flagellar propulsion for colonial integrity.[154] The germline-soma separation further incurs repair costs, prioritizing germline integrity over somatic maintenance to ensure reproductive fidelity across generations.[155] Representative examples illustrate varying degrees of multicellular complexity. Sponges (Porifera), emerging over 600 million years ago, represent simple multicellularity without true tissues, relying on choanoflagellate-derived adhesion for larval settlement and cell aggregation into functional modules like choanocytes for feeding.[156] In contrast, cnidarians, diverging around 580 million years ago, achieved tissue-level organization with epithelial layers and nerve nets, utilizing Wnt signaling for polarity and minicollagens for connective tissues that support contractility and predation.[157] These lineages highlight how incremental adhesion and signaling innovations built upon unicellular precursors to foster multicellular advantages in diverse environments.Evolutionary Origins
Origin of Life and First Cells
The origin of life on Earth is thought to have arisen through a process of chemical evolution, where simple inorganic molecules gradually formed complex organic compounds under prebiotic conditions. This abiogenesis likely occurred in a reducing atmosphere or aquatic environments rich in energy sources, leading to the synthesis of life's building blocks such as amino acids, nucleotides, and lipids. Key experiments and hypotheses have illuminated these early stages, emphasizing the transition from non-living chemistry to self-sustaining systems capable of replication and metabolism.[158] A landmark demonstration of chemical evolution came from the Miller-Urey experiment conducted in 1953, which simulated early Earth conditions by mixing gases like methane, ammonia, hydrogen, and water vapor in a closed system and subjecting them to electrical sparks to mimic lightning. After one week, the reaction produced several amino acids, including glycine and alanine, at yields up to 2-5% of the initial carbon input, showing that organic monomers essential for proteins could form abiotically from inorganic precursors. Subsequent analyses and variations of the experiment, using revised atmospheric compositions, have confirmed the synthesis of over 20 amino acids under diverse conditions, underscoring the plausibility of prebiotic organic synthesis.[159][160] Central to many theories of life's emergence is the debate between "replication first" and "metabolism first" scenarios, which address the primacy of genetic information or energy-processing networks in proto-life. The replication-first view, embodied by the RNA world hypothesis, posits that RNA molecules served dual roles as both genetic material and catalysts in early evolution, predating DNA and proteins. Proposed in 1986, this idea gained support from the discovery of ribozymes—RNA enzymes capable of self-splicing and peptide bond formation—and laboratory demonstrations of RNA polymerization from prebiotic nucleotides, suggesting RNA could have enabled the first self-replicating systems around 4 billion years ago. In contrast, the metabolism-first hypothesis argues that autocatalytic cycles of chemical reactions, such as those involving iron-sulfur minerals, established energy gradients and metabolic pathways before replicators evolved, as evidenced by simulations showing sustained geochemical cycles in hydrothermal settings that produce organic intermediates without templated replication.[158][161][162] The formation of protocells—primitive, membrane-bound compartments—represents a critical step toward cellular organization, encapsulating reactive molecules to concentrate chemistry and enable division. Lipid vesicles, formed spontaneously from amphiphilic fatty acids in aqueous environments, can grow by incorporating new lipids and divide through shear forces or osmotic pressure, mimicking basic cellular behaviors; experiments show these vesicles encapsulate RNA and maintain internal pH gradients for up to hours. Coacervates, liquid droplets arising from phase separation of polyelectrolytes like peptides and nucleic acids, provide an alternative model, as they concentrate biomolecules without lipids and exhibit selective permeability, with recent studies demonstrating their stability in saline conditions relevant to early oceans. Hydrothermal vents, particularly alkaline ones on the seafloor, are favored sites for protocell assembly due to their thermal gradients and mineral-rich fluids, where lipid membranes form at the interface between vent alkaline solutions and acidic seawater, facilitating proton gradients akin to those in modern cells.[163][164][165] Geological evidence places the emergence of the first cells between 4.1 and 3.8 billion years ago, shortly after Earth's oceans formed around 4.4 billion years ago, with the oldest undisputed signs of microbial life in 3.5-billion-year-old stromatolites from Western Australia. These layered structures, formed by cyanobacterial mats trapping sediments, indicate photosynthetic prokaryotes thrived by 3.48 billion years ago, as confirmed by isotopic analysis showing carbon fractionation consistent with biological activity. Microfossils and chemical biomarkers from 3.7-billion-year-old rocks in Greenland further support this timeline, though debates persist over abiotic mimics. In November 2025, AI analysis revealed chemical biosignatures of life in 3.3-billion-year-old rocks and evidence of oxygenic photosynthesis in rocks up to 2.5 billion years old, further supporting early microbial activity.[166][167][168] Recent advances in prebiotic chemistry, accelerated by machine learning simulations as of 2025, have refined models of these early processes by predicting reaction pathways and molecular stabilities with high accuracy. For instance, machine learning-based simulations have explored prebiotic reactions like nucleotide phosphoester bond formation, enabling microsecond-scale studies with near-quantum accuracy at reduced computational cost.[169] These computational tools bridge gaps in experimental data, supporting hybrid metabolism-replication models and highlighting the role of mineral surfaces in catalyzing both pathways.Emergence of Eukaryotic Cells
The emergence of eukaryotic cells represents a pivotal evolutionary transition from prokaryotic ancestors, primarily driven by endosymbiotic events that integrated bacterial symbionts into a host cell, leading to the development of complex organelles and cellular structures.[170] The foundational endosymbiosis involved the engulfment of an alpha-proteobacterium by an archaeal-like host cell, giving rise to mitochondria approximately 1.8 to 2.2 billion years ago (Ga).[171] This event provided the host with enhanced energy production through oxidative phosphorylation, fundamentally altering cellular metabolism and enabling larger cell sizes and increased complexity.[172] Subsequently, in the lineage leading to photosynthetic eukaryotes, a primary endosymbiosis occurred around 1 to 1.5 Ga, when a cyanobacterium was incorporated as the progenitor of chloroplasts in the ancestor of Archaeplastida.[173] Secondary endosymbioses, involving the engulfment of primary plastid-bearing algae by non-photosynthetic eukaryotes, further diversified plastid types in groups such as chromalveolates, occurring as recently as 60 million years ago in some cases.[174] Compelling evidence for these endosymbiotic origins includes the retention of distinct genetic and structural features in mitochondria and chloroplasts that mirror their bacterial ancestors. Mitochondria possess their own circular genome (mtDNA), encoding 13 to 37 genes primarily involved in respiration and protein synthesis, a reduced set compared to free-living bacteria but indicative of an independent prokaryotic heritage.[171] Both organelles are bounded by double membranes, with the inner membrane derived from the symbiont's plasma membrane and the outer from the host's phagocytic vesicle.[175] Additionally, their ribosomes are of the 70S type, characteristic of bacteria, contrasting with the 80S ribosomes of the eukaryotic cytosol. Recent models, supported by phylogenetic reconstructions updated around 2023, propose that peroxisomes arose from the endoplasmic reticulum (ER), possibly linked metabolically to the mitochondrial lineage, enhancing compartmentalization of oxidative processes.[176] Beyond endosymbiosis, key innovations distinguished early eukaryotes, including the evolution of the nuclear envelope, likely formed by invaginations and reshaping of the ER to enclose the genome, facilitating regulated gene expression around 1.5 Ga.[177] The cytoskeleton emerged through the co-option and diversification of prokaryotic filament systems, incorporating actin, tubulin, and intermediate filaments for dynamic intracellular transport, cell division, and shape maintenance, with roots in the archaeal host.[178] Sexual reproduction also arose as an early eukaryotic trait, enabling genetic recombination via meiosis and cell fusion, likely evolving once in the last eukaryotic common ancestor (LECA) to repair DNA damage and promote diversity.[179] The LECA, estimated to have existed around 1.8 Ga, encompassed these features and marked the diversification of eukaryotic lineages.[180] Fossil evidence supports this timeline, with Grypania spiralis, interpreted as an early eukaryotic alga, dating to approximately 1.87 Ga in Paleoproterozoic strata. These developments collectively enabled eukaryotes to exploit new ecological niches, setting the stage for multicellularity.References
- https://bio.libretexts.org/Bookshelves/[Microbiology](/page/Microbiology)/Microbiology_(OpenStax)/03:_The_Cell/3.02:_Foundations_of_Modern_Cell_Theory




